EvoLaps is a user-friendly web application designed to visualize the spatial and temporal spread of pathogens.
It takes an annotated tree as input, such as a maximum clade credibility tree obtained through
phylogeographic inference.
By following a top-down reading of the tree, transitions (latitude/longitude changes from a node to its children)
are represented on a cartographic background using graphical paths.
The complete set of paths forms the phylogeographic scenario.
A raw reading of these transitions can produce complex scenarios. EvoLaps helps analyze them with:
- 2D/3D visualization of scenarios with multiple layouts and extensive style customization,
- multiple graphical variables with time-dependent gradients for enhanced path display,
- cross-highlighting and selection between the phylogenetic tree and the phylogeographic scenario,
- clustering of localities into spatial clusters to generate synthetic views,
- animation of the phylogeographic scenario using phylogenetic tree brushing,
- support for both discrete and continuous model analyses,
- a customizable geographic map with support for environmental data layers,
- interactive phylogenetic tree editing,
|
EvoLaps video showcase
Contact
-
francois.chevenet@ird.fr if you would like to use EvoLaps and see it evolve with the integration of new features (modeling in epidemiology, phylogenetics, phylodynamics), feel free to contact me.
Funding
- ANRS-24-PEPR MIE-0003 PEPR MIE 2024. PReViX project (Pandemic preparedness to Respiratory Virus X)
- Agence Nationale pour la Recherche through the grant GENOSPACE ANR-16-CE02-0008.
- PALADIN project, publicly funded through the French National Research Agency under the “Investissements d’avenir” program with the reference ANR-10-LABX-04-01 Labex CEMEB.
Warning
- 'Shift' load EvoLaps to refresh the cache of the navigator. Hold down the 'Shift' key while clicking the reload button, or 'command' key (or CTRL key) + R
API
Citation
- EvoLaps version 3 - Chevenet F, Vitré T, Cadar D, Fichet-Calvet E, Fargette D, Bastide P, Guindon S. EvoLaps 3: Next-Level phylogeographic visualization.
under construction
- EvoLaps version 2 - Chevenet F, Fargette D, Bastide P, Vitré T, Guindon S. EvoLaps 2: Advanced phylogeographic visualization. Virus Evol. 1 janv
2024;10(1):vead078.

- EvoLaps version 1 - Chevenet F, Fargette D, Guindon S, Bañuls AL. EvoLaps: a web interface to visualize continuous phylogeographic recon-
structions. BMC Bioinformatics. 27 sept 2021;22(1):463.

Thanks
- D. Hubert for system administration
- A. Prudhon for English editing
|
|
|
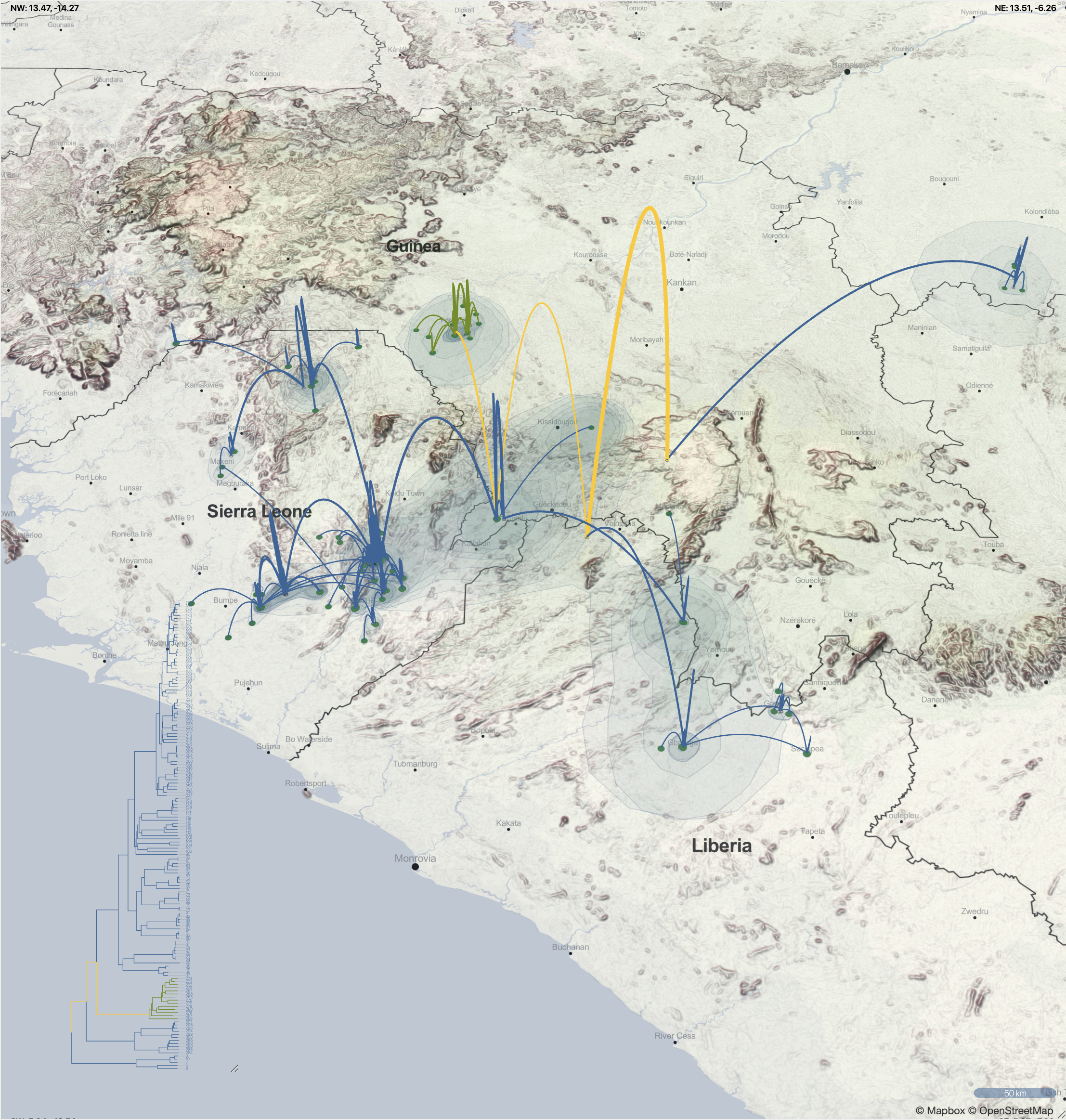
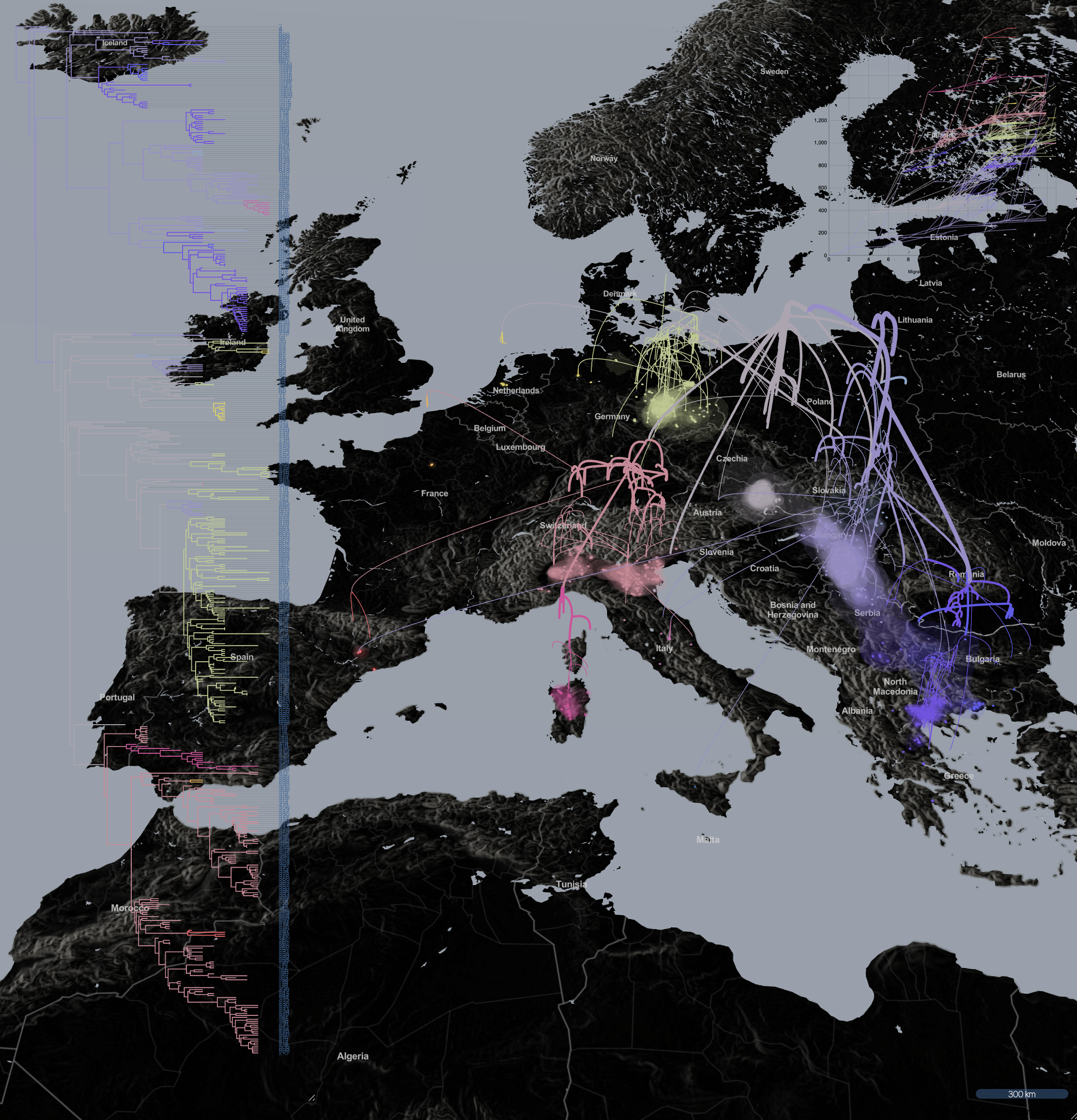
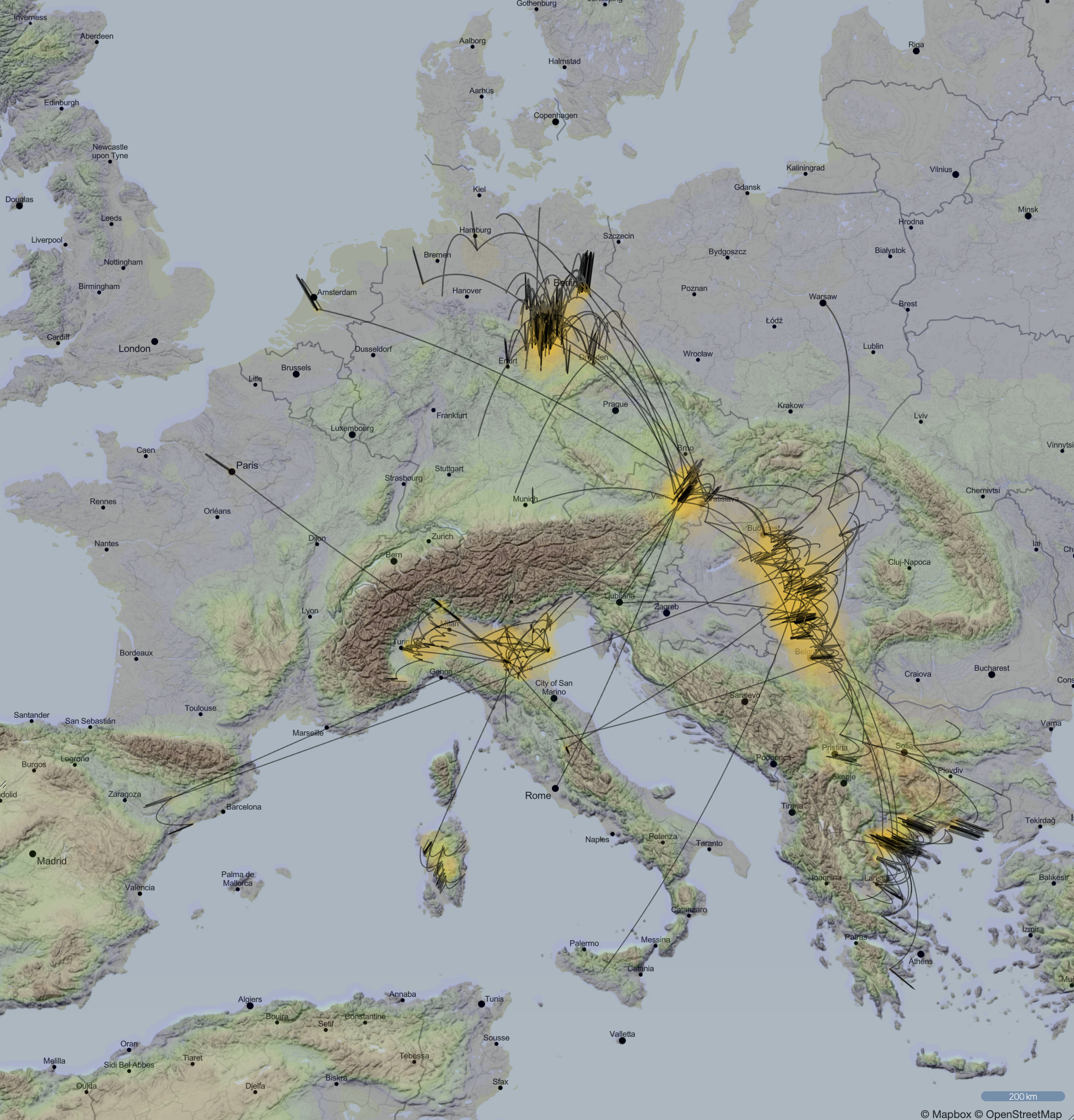
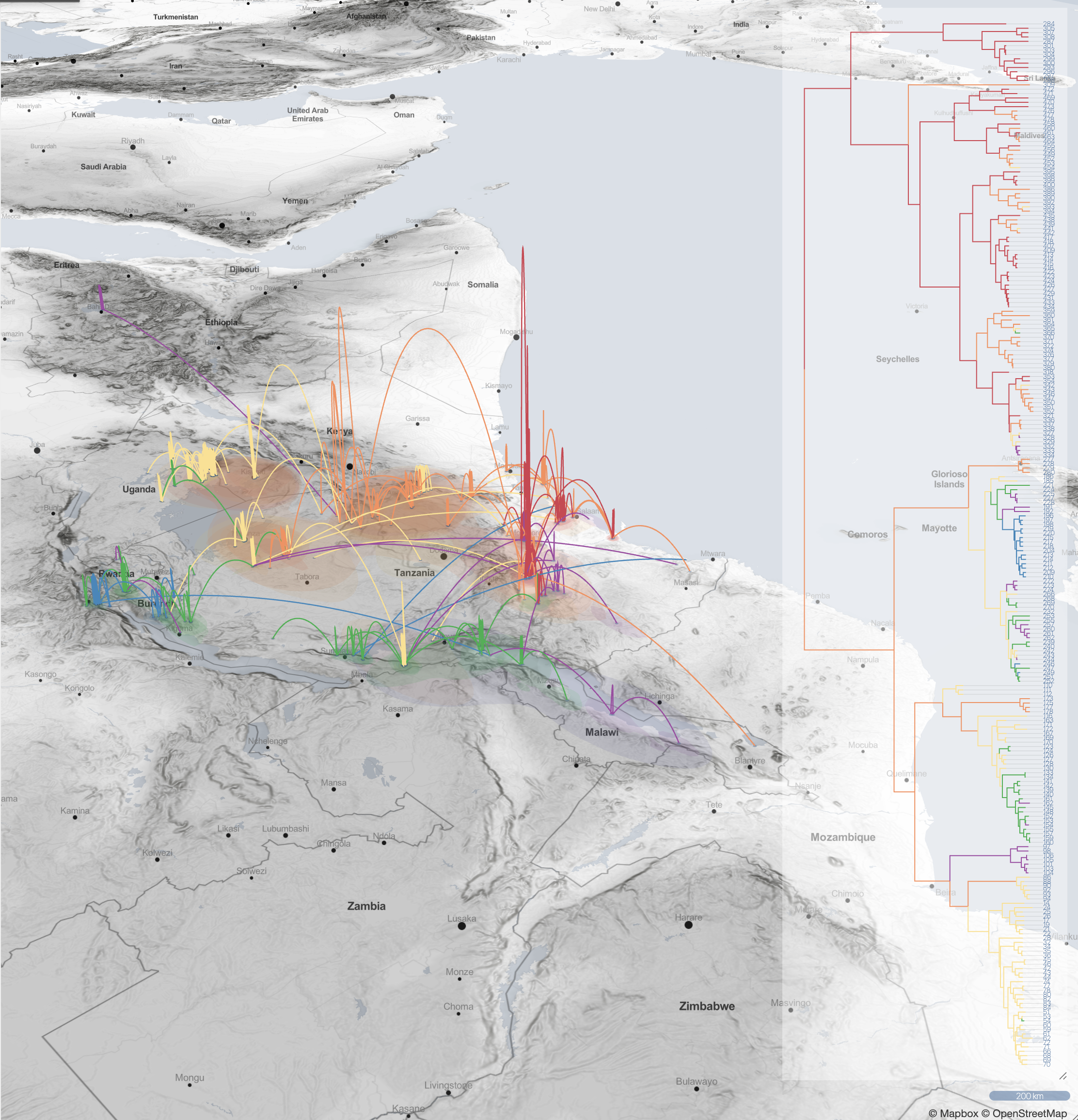
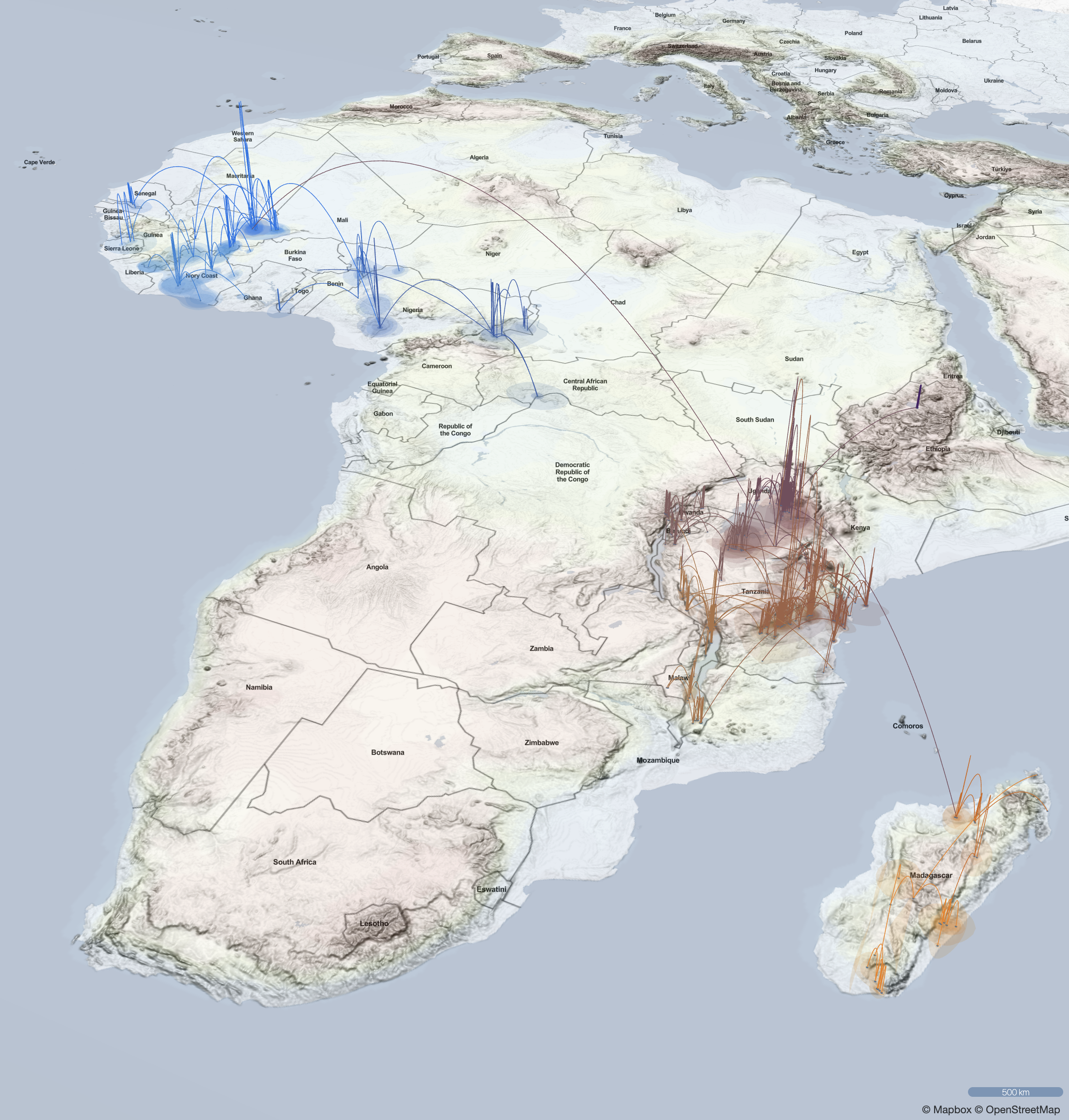







Spatio-temporal spread of Lassa virus and a new rodent host in the Mano River Union area, West Africa. (Bangura et al., 2024)