|
1
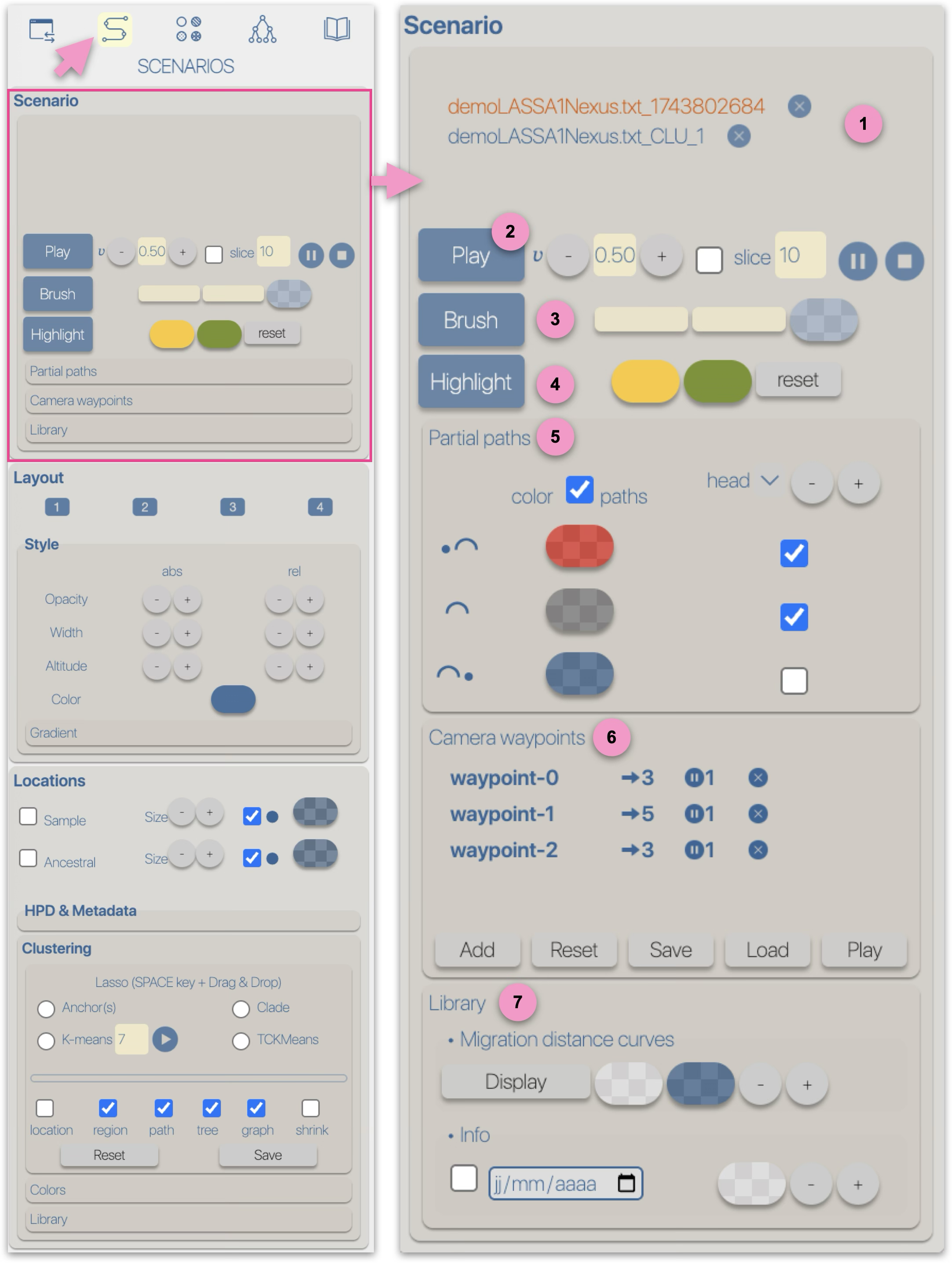
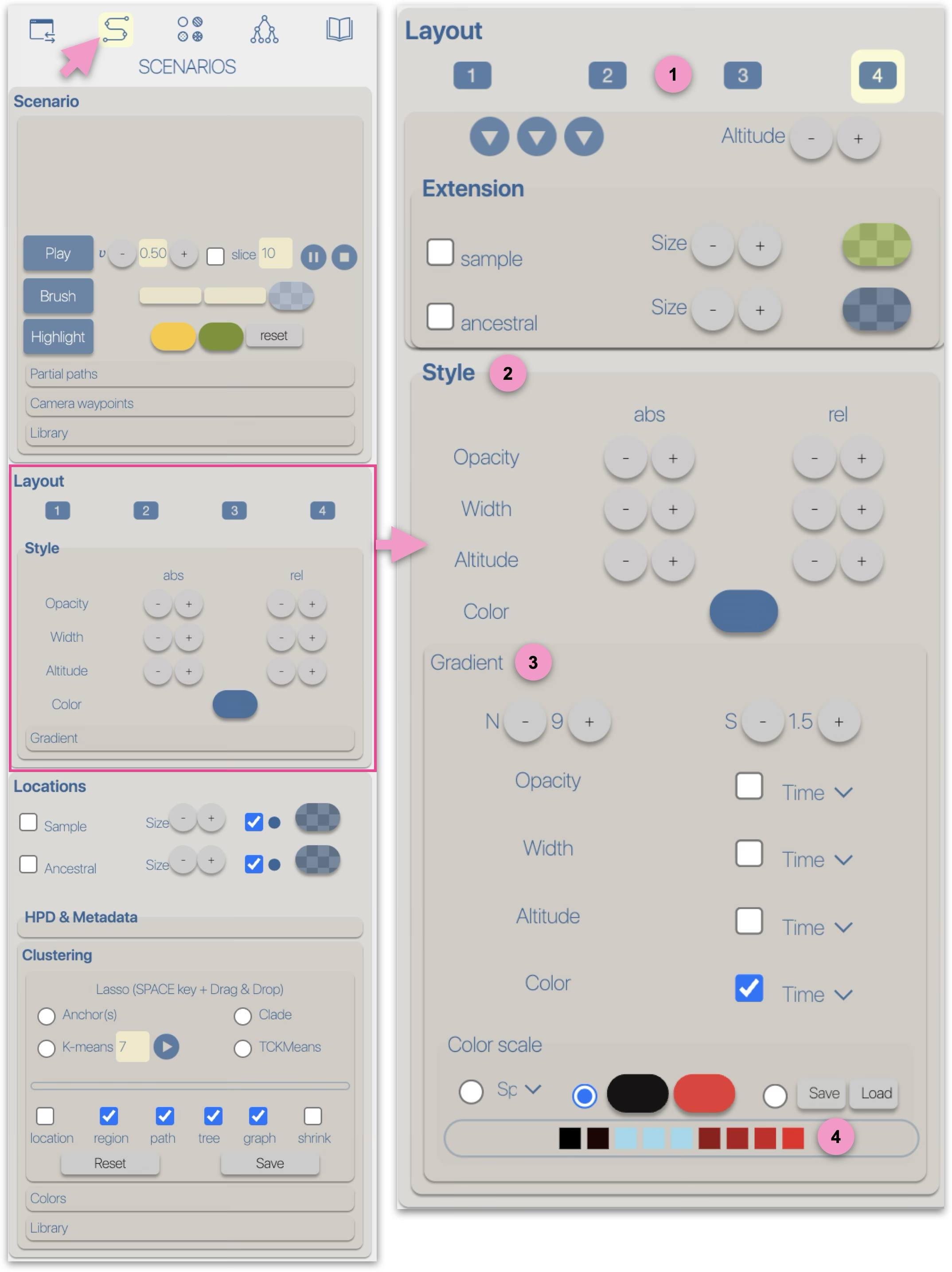
The graphical representations of phylogeographic scenarios (layouts) are grouped
into families (currently four), based on the methods used to compute a path connecting points (latitude/longitude)
associated with nodes of the phylogenetic tree (a parent node to its child nodes).
For example, and without going into too much technical detail, layout family 1 implements
a Bézier curve smoothing technique using three control points:
- a first point (latitude/longitude) associated with a parent node,
- a second point (latitude/longitude) associated with a child node,
- an intermediate point calculated as the average of the latitude and longitude of the two previous points.
Layout family 2 also uses a smoothing method based on 3 control points: the points associated with the parent and child nodes,
but the intermediate point lies on a circle of radius *r* and at an angle *alpha*, with the circle centered
on the average latitude and longitude of the parent/child points.
Layout family 3 uses a smoothing method based on 4 control points: the points associated with the parent and child nodes,
and two intermediate points, each located on a circle of radius *r* and angle *alpha*, centered
respectively on the parent and the child node.
Due to these different computation methods, each layout family is associated with specific parameters/controls.
For example, family 1 provides an offset control for the latitude and longitude of the intermediate point.
Family 2 offers offset controls for the radius and angle of the circle carrying the intermediate point.
Other layout-family-specific parameters allow for visual variations of the phylogeographic scenarios.
For instance, family 4, which introduces a notion of altitude, provides variants based on the path shape: linear, curved, or rectangular,
as well as an altitude offset and the drawing of the vertical projection of the points onto the surface.
Nota Bene:
the graphical rendering quality of phylogeographic scenarios varies depending on
the different families/methods used to compute the paths, and may be more or less suitable depending on
the nature of the dataset. It is recommended to test them.
2
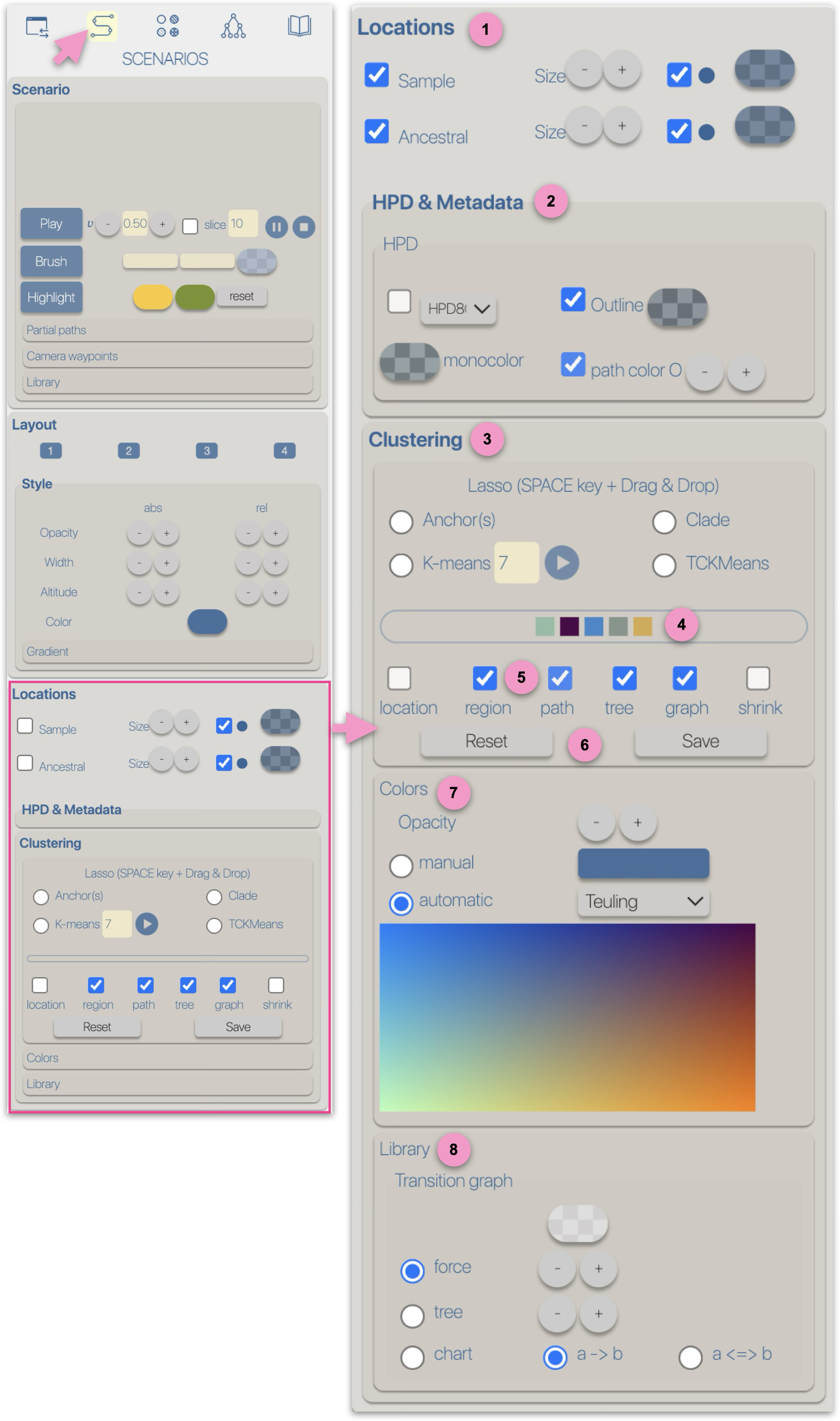
Regardless of the layout used to represent the phylogeographic scenario, it can be modified using
four graphical variables: opacity, stroke thickness, altitude (curvature), and color.
Apart from color, modifications (increase/decrease) can be either 'absolute' (abs): the same value is applied to all paths in the scenario, or
relative (rel): the existing value of each path is incremented (or decremented) individually.
These modifications can be performed manually (using -/+ buttons) or automatically by applying a gradient.
3
Gradient Opacity, stroke thickness, altitude (curvature), and color can be associated with a gradient.
This gradient is configurable:
- N is the number of classes
- S is a stringency parameter between classes — the higher the S value, the stronger the contrast between classes
A gradient can be based on:
- Time: this gradient is based on the cumulative branch length from the node to the root of the phylogenetic tree
- Distance: this gradient is computed as the sum of geographical distances along the path from the node to the tree root (bottom-up recursive process).
It is the sum of orthodromic distances (e.g., great-circle distances based on latitude and longitude between two points on a sphere)
- Speed: this gradient is based on the Distance/Time ratio of the two previous gradients
The color gradient has three modes:
- using a color scale. The colors in a scale are optimized, and the choice of scales is restricted depending on the number of gradient classes
- selecting a start and end color
- saving/loading a list of specific colors.
The colors must be in hexadecimal format without transparency, listed in square brackets, for example:
["#d53e4f", "#fc8d59", "#fee08b", "#b5e61d", "#22b14b", "#a349a4", "#3288bd"].
In this example, the gradient can use up to 7 classes. The colors will be used in the order they appear in the list.
It is possible to use multiple gradients simultaneously — for example, a stroke thickness gradient based on time and a color gradient based on distance.
4
The color gradient classes (associated with time, distance, or speed) are represented by a list of colored boxes.
Clicking on a box allows you to change the color of the corresponding class.
Assigning the same color to multiple adjacent classes enables a non-linear color gradient.
|
|

 Map,Tree, Library and Manual button : display ON/OFF the corresponding window and bring it to the foreground
Map,Tree, Library and Manual button : display ON/OFF the corresponding window and bring it to the foreground MapBox: on/off display of the MapBox/OpenStreetMap copyrights
MapBox: on/off display of the MapBox/OpenStreetMap copyrights Tooltip: on/off display of information linked to the controls of the interface (to the right of the EvoLaps logo)
Tooltip: on/off display of information linked to the controls of the interface (to the right of the EvoLaps logo) .
. .
.