|
EvoLaps URL: www.evolaps.org.
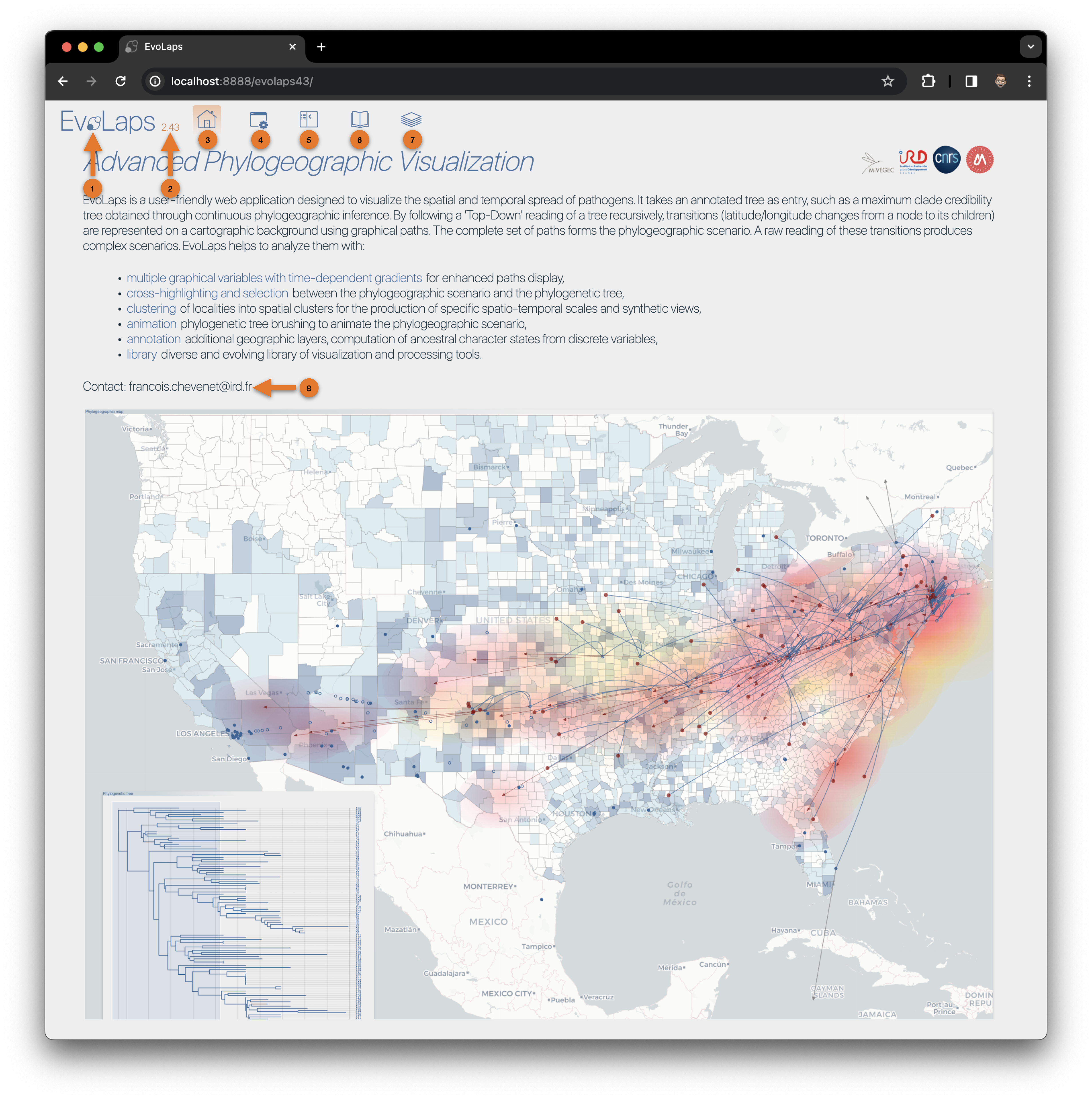
(1) EvoLaps icon, animation when communication with the server side of EvoLaps
(3) EvoLaps current version
(3) EvoLaps Home
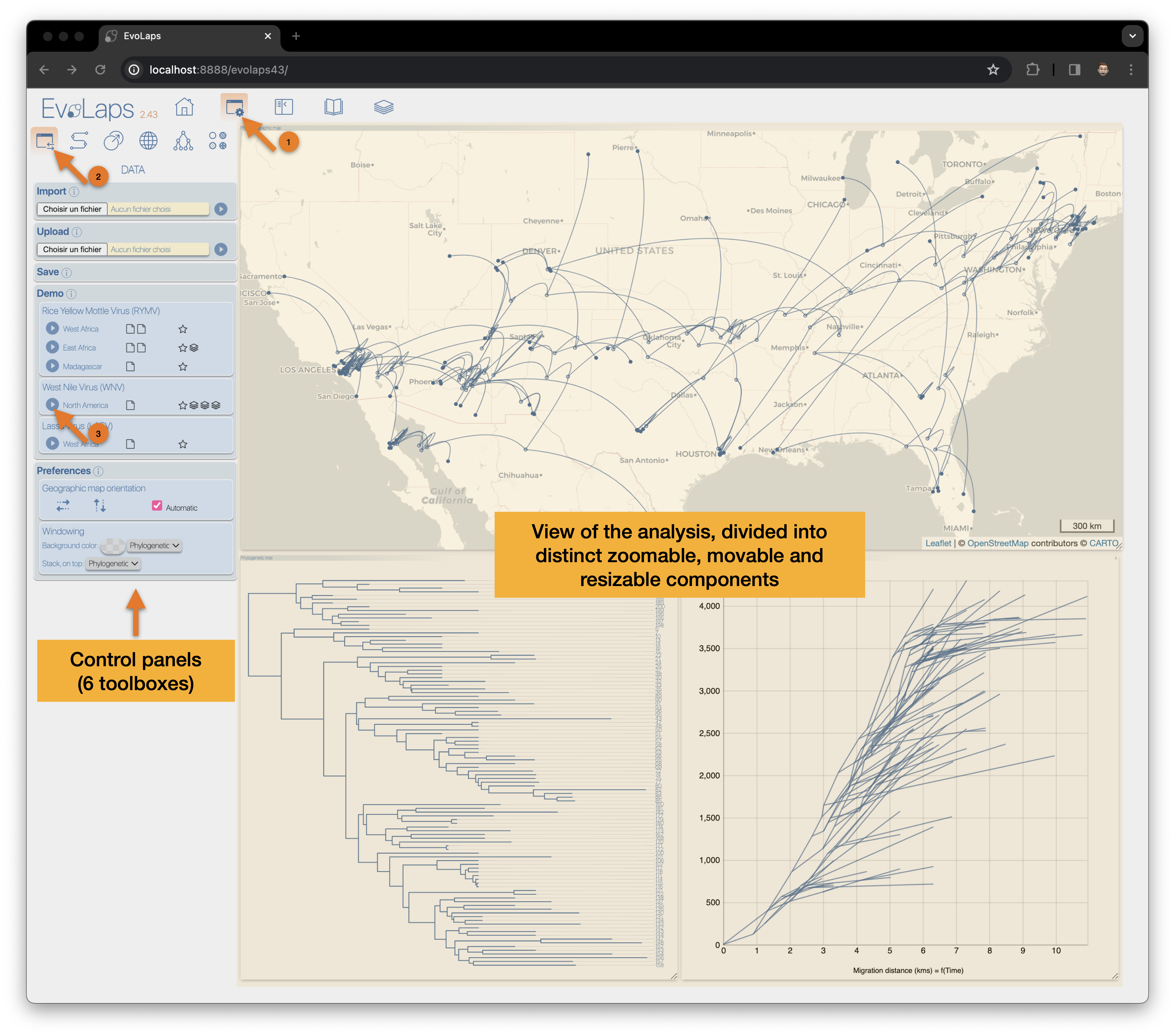
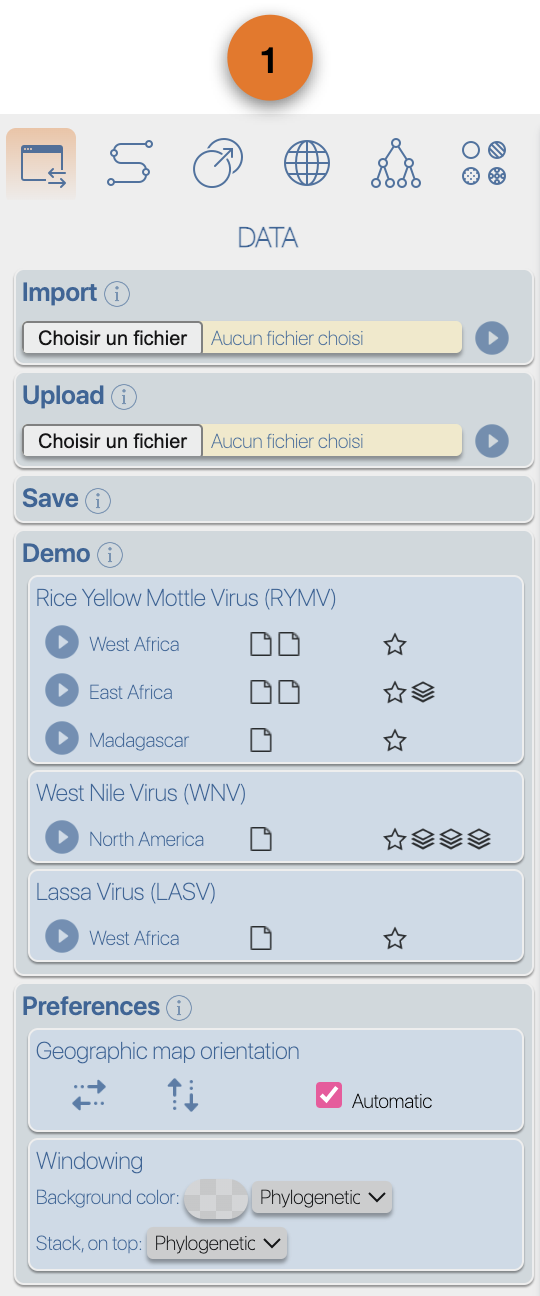
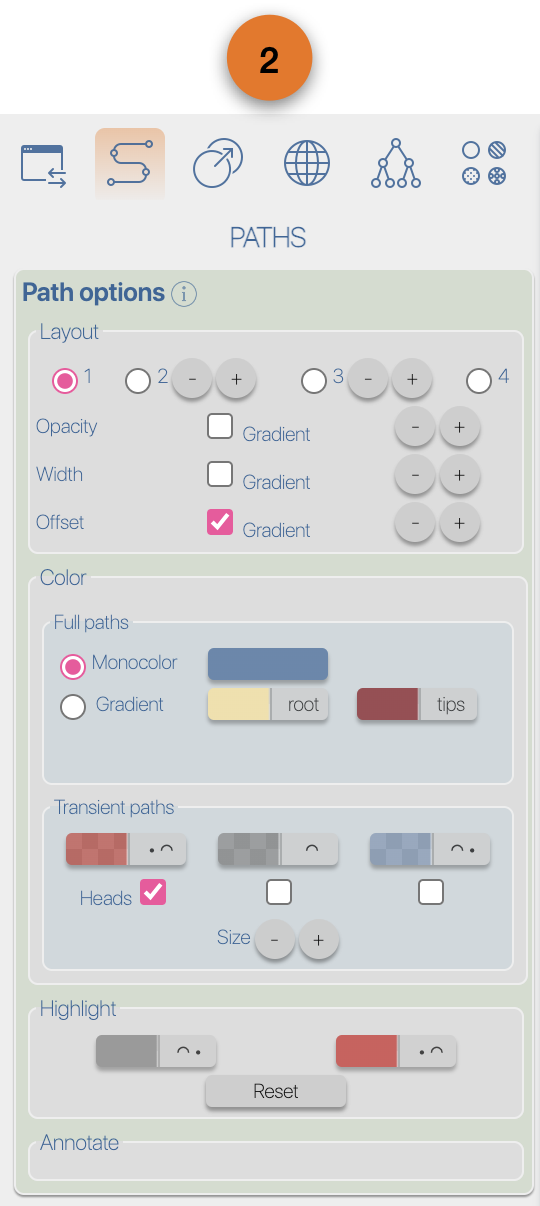
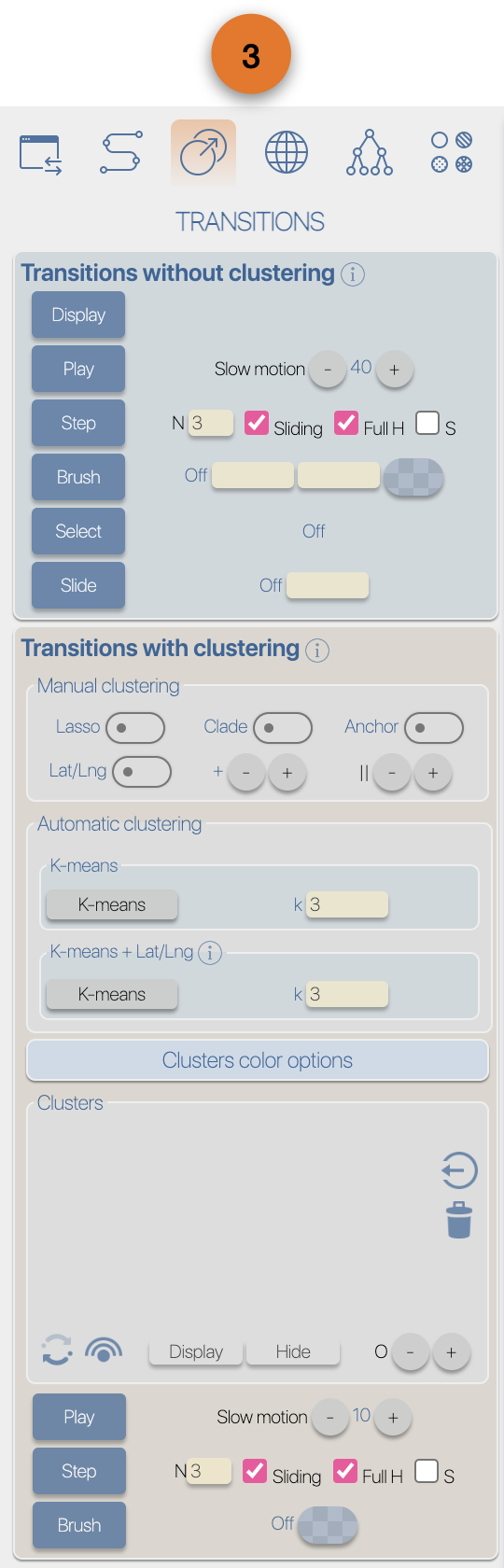
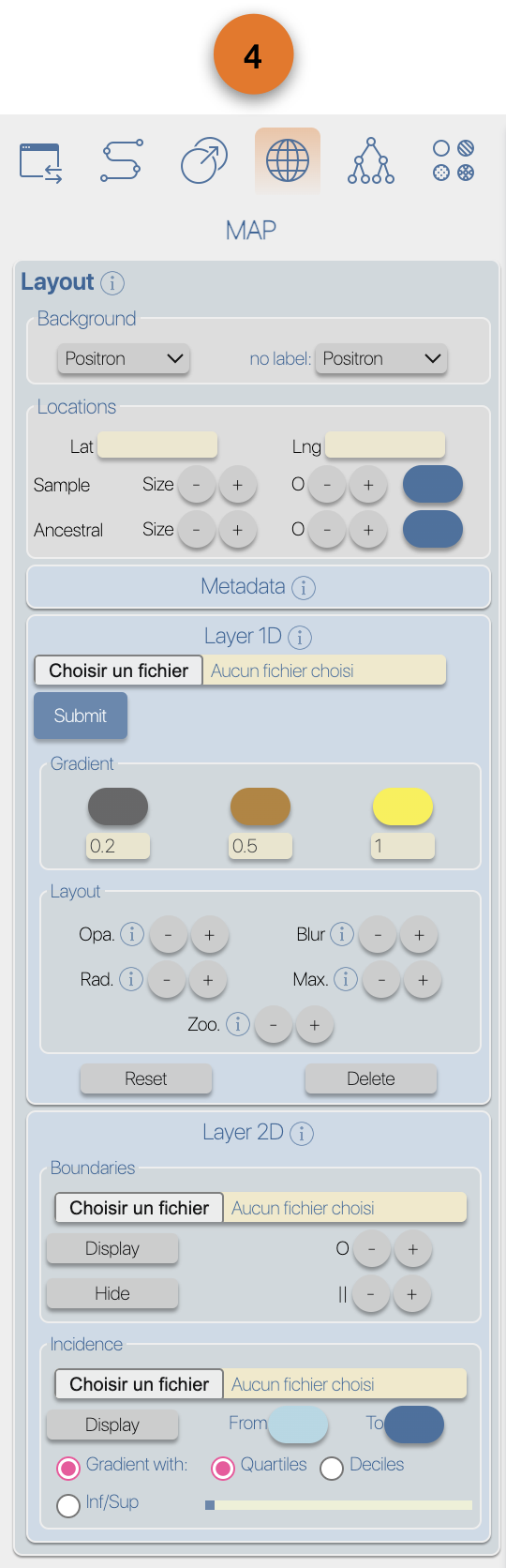
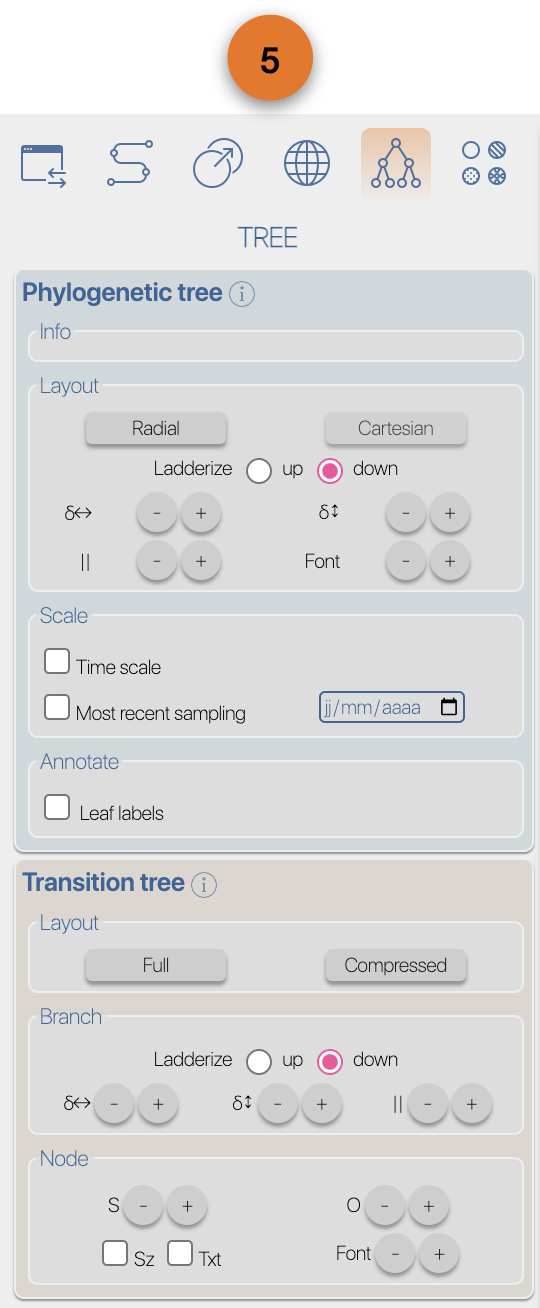
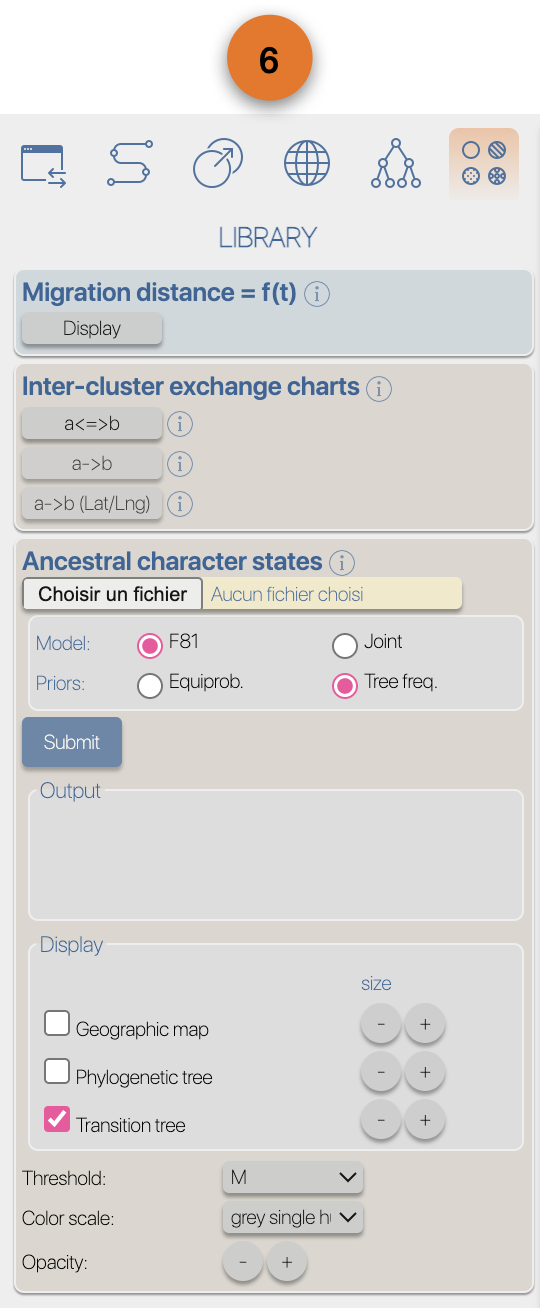
(4) EvoLaps toolboxes for analyses
(5) Display/hide toolboxes for more space before screen captures and so on
(6) EvoLaps documentation. It can be accessed in a different navigator window at the following URL
www.evolaps.org/evolapsdocs
(7) EvoLaps News
(8) Do not hesitate to contact me (francois.chevenet@ird.fr) if you need help (data importation...), detect any bugs or if you want to see additional functionalities to be integrated into EvoLaps.
|
|