|
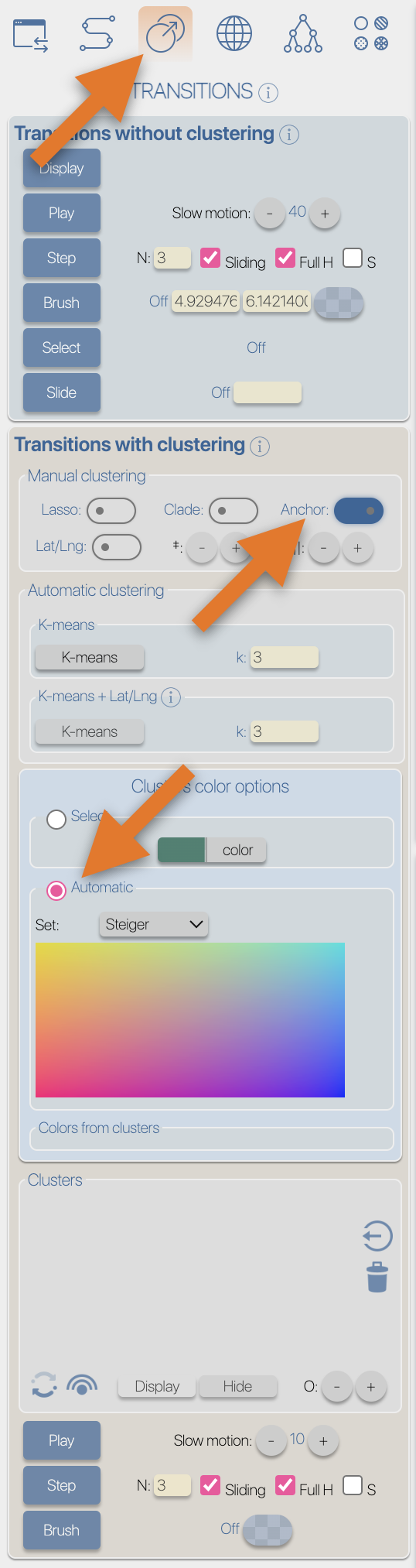
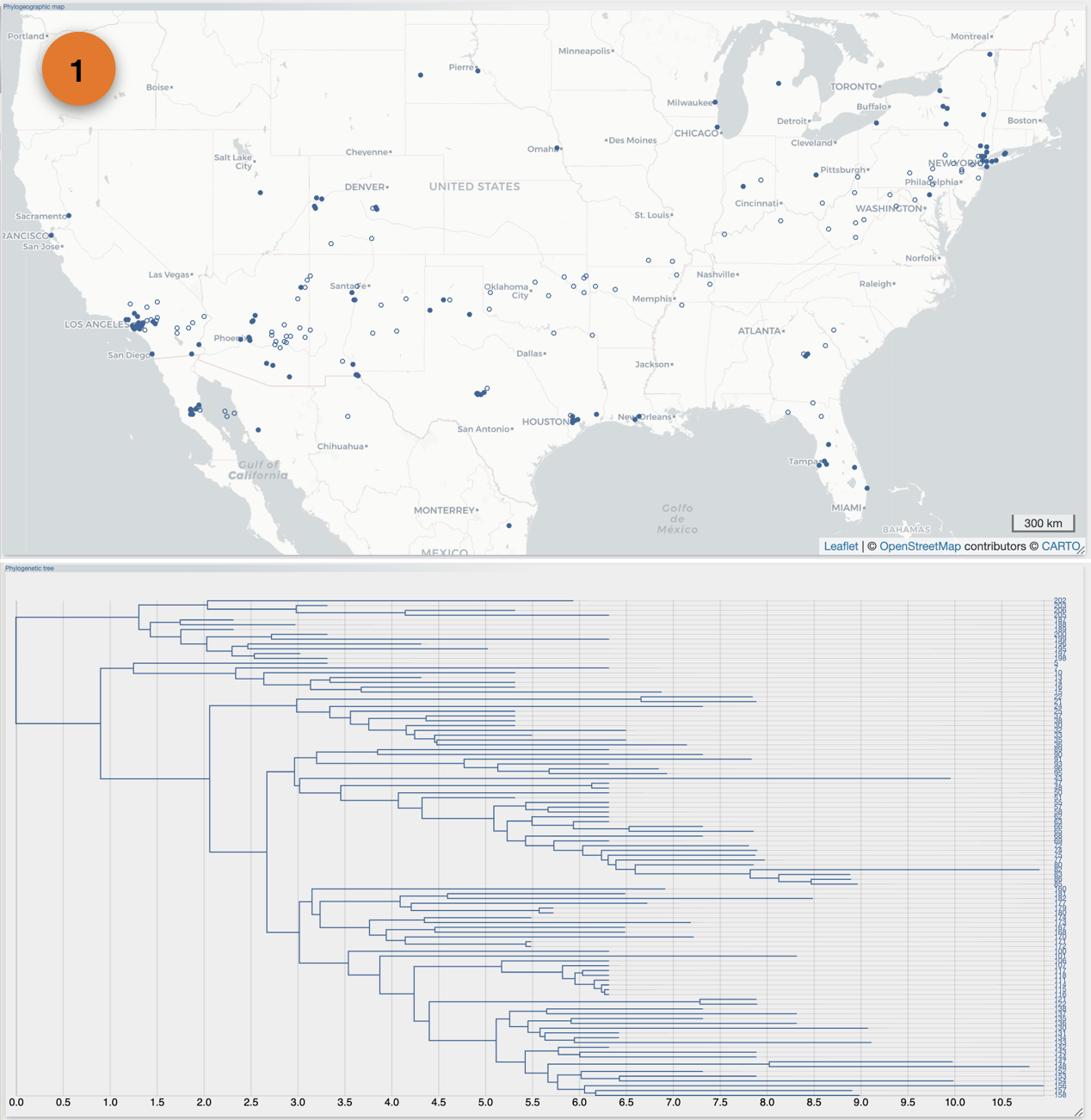
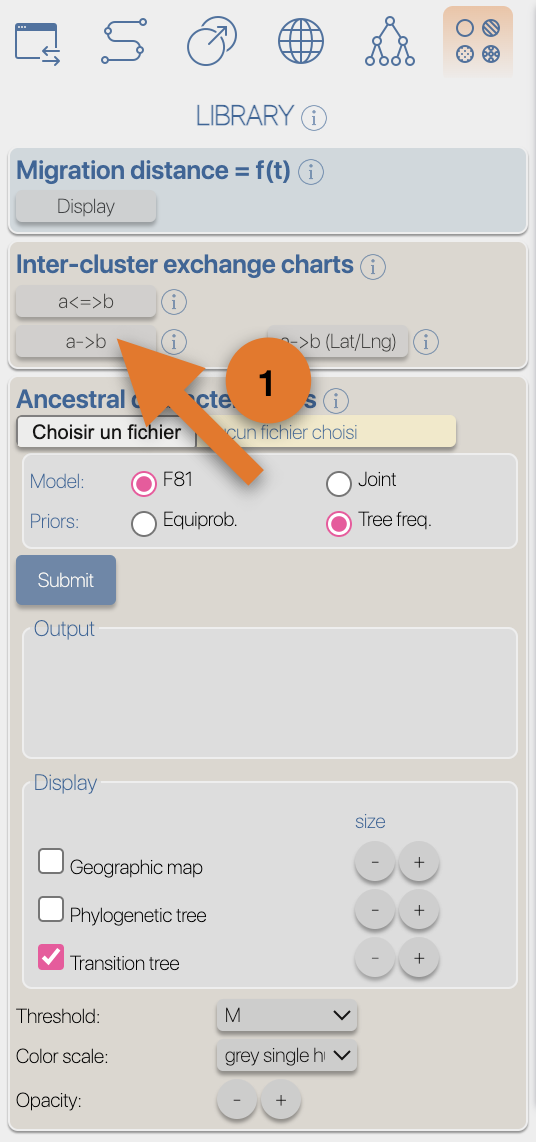
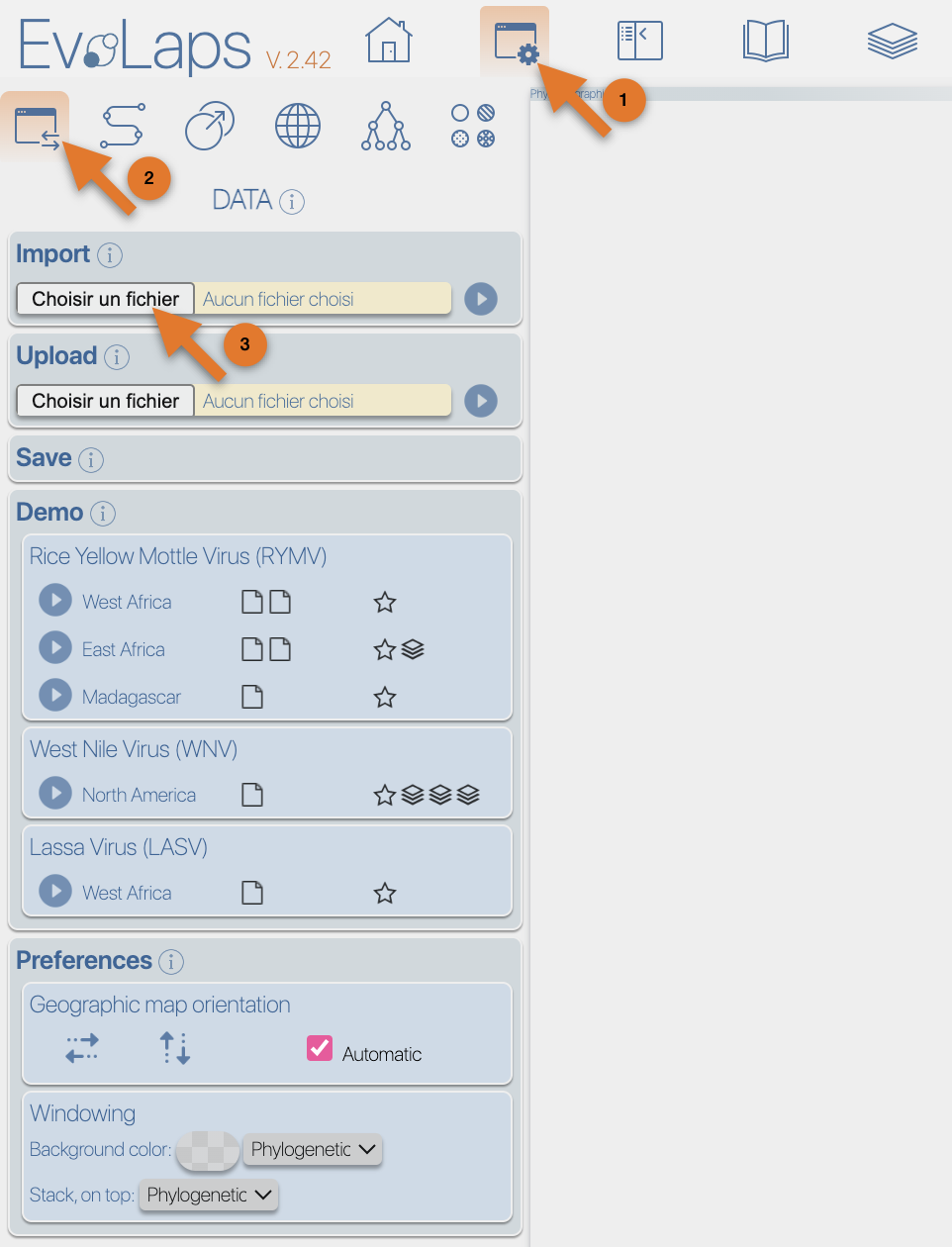
From the Top toolbox, select the "Process" button (1), select the 'DATA' toolbox (2), click the 'select file' button from the 'import' section (3), select the input file then click the 'submit' button.
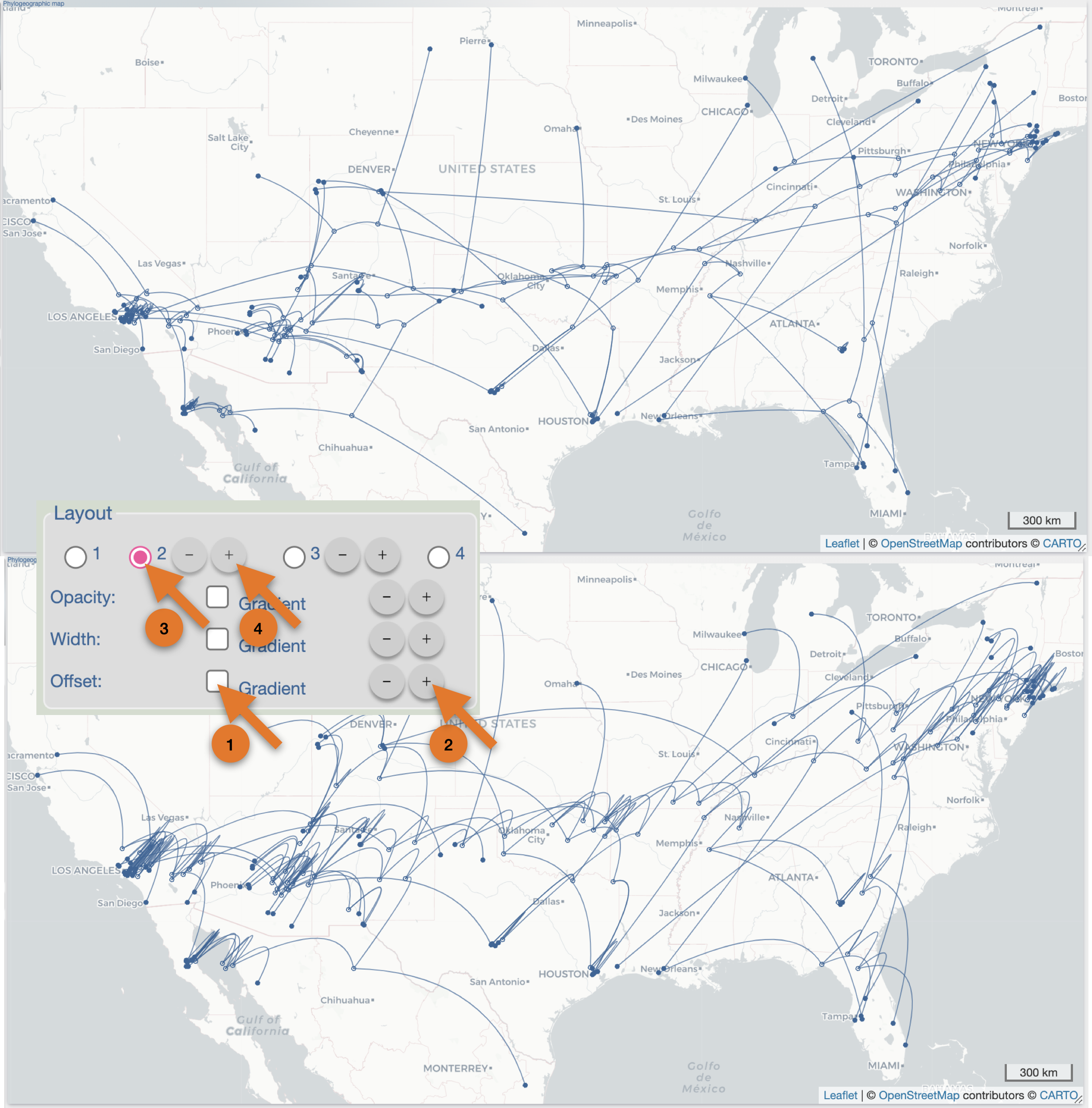
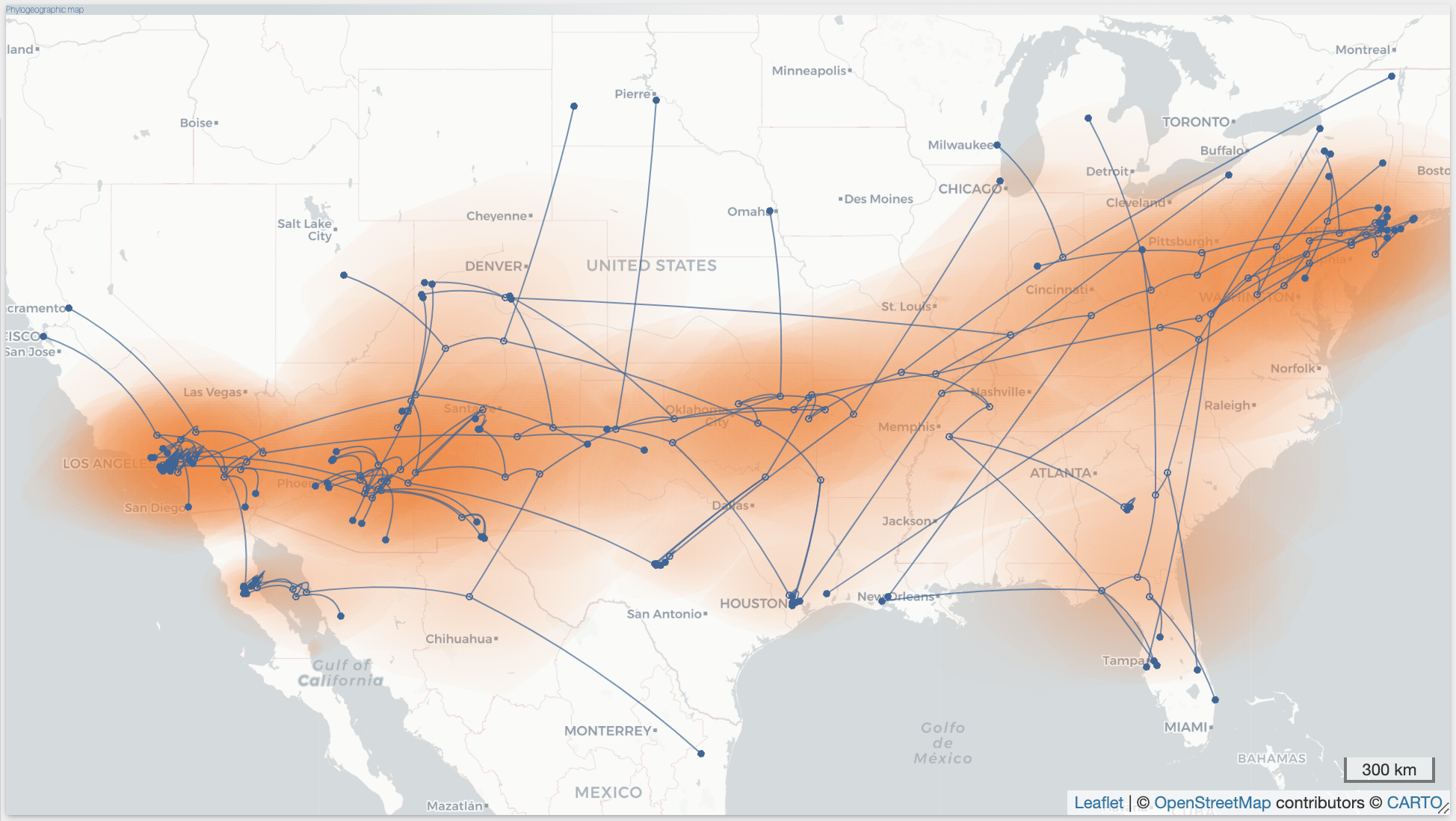
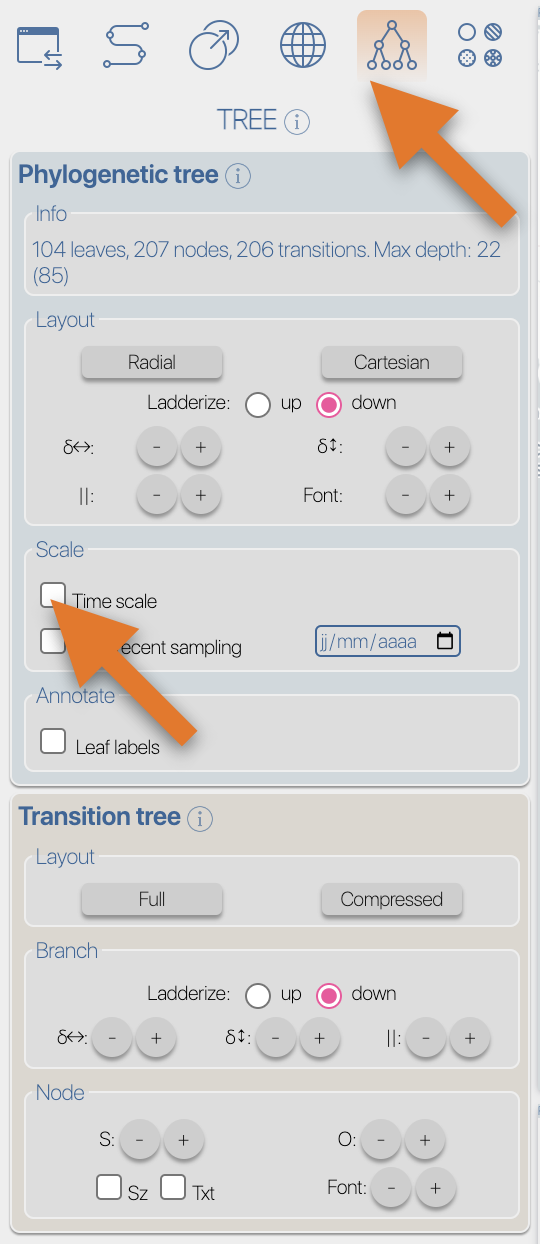
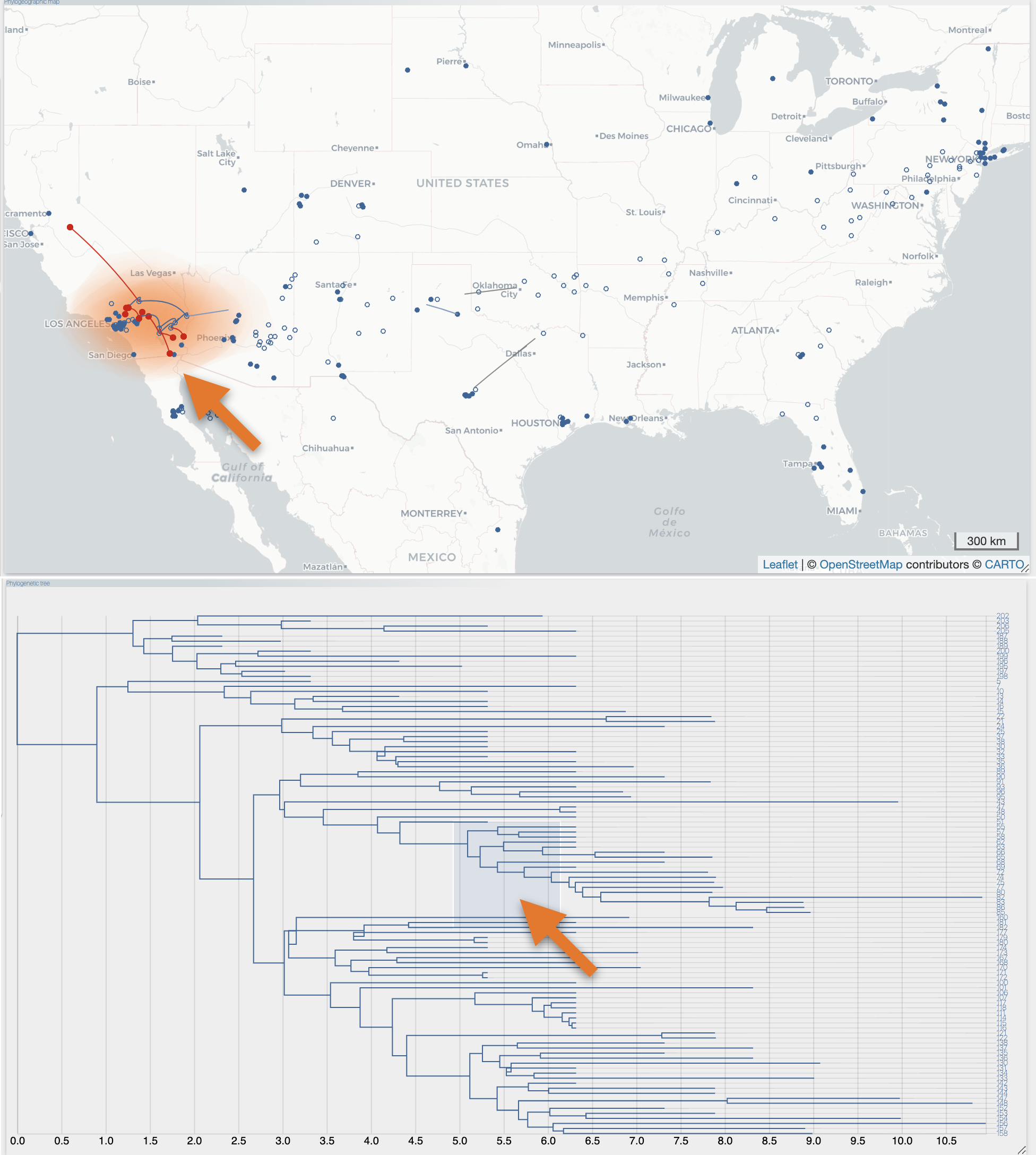
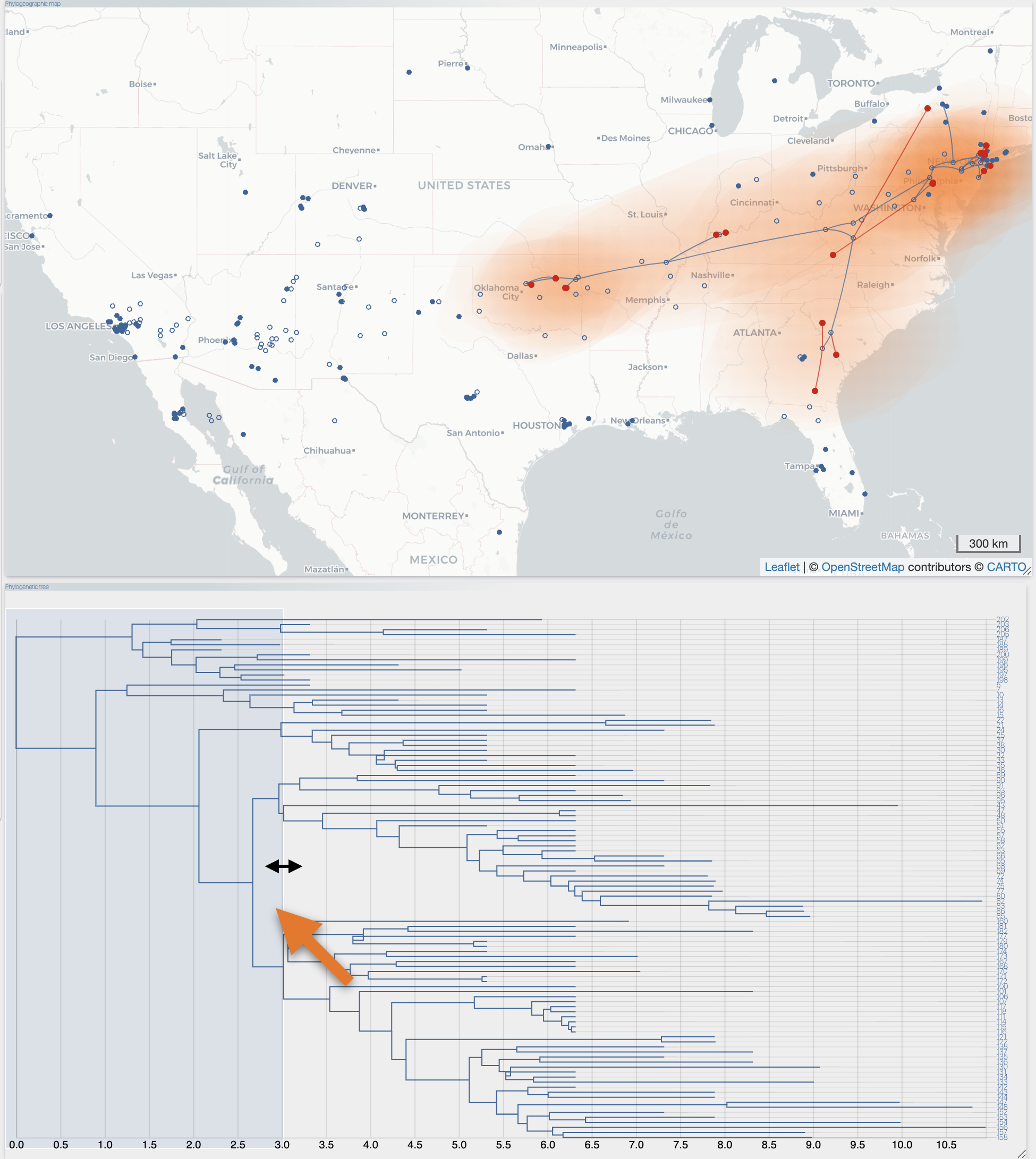
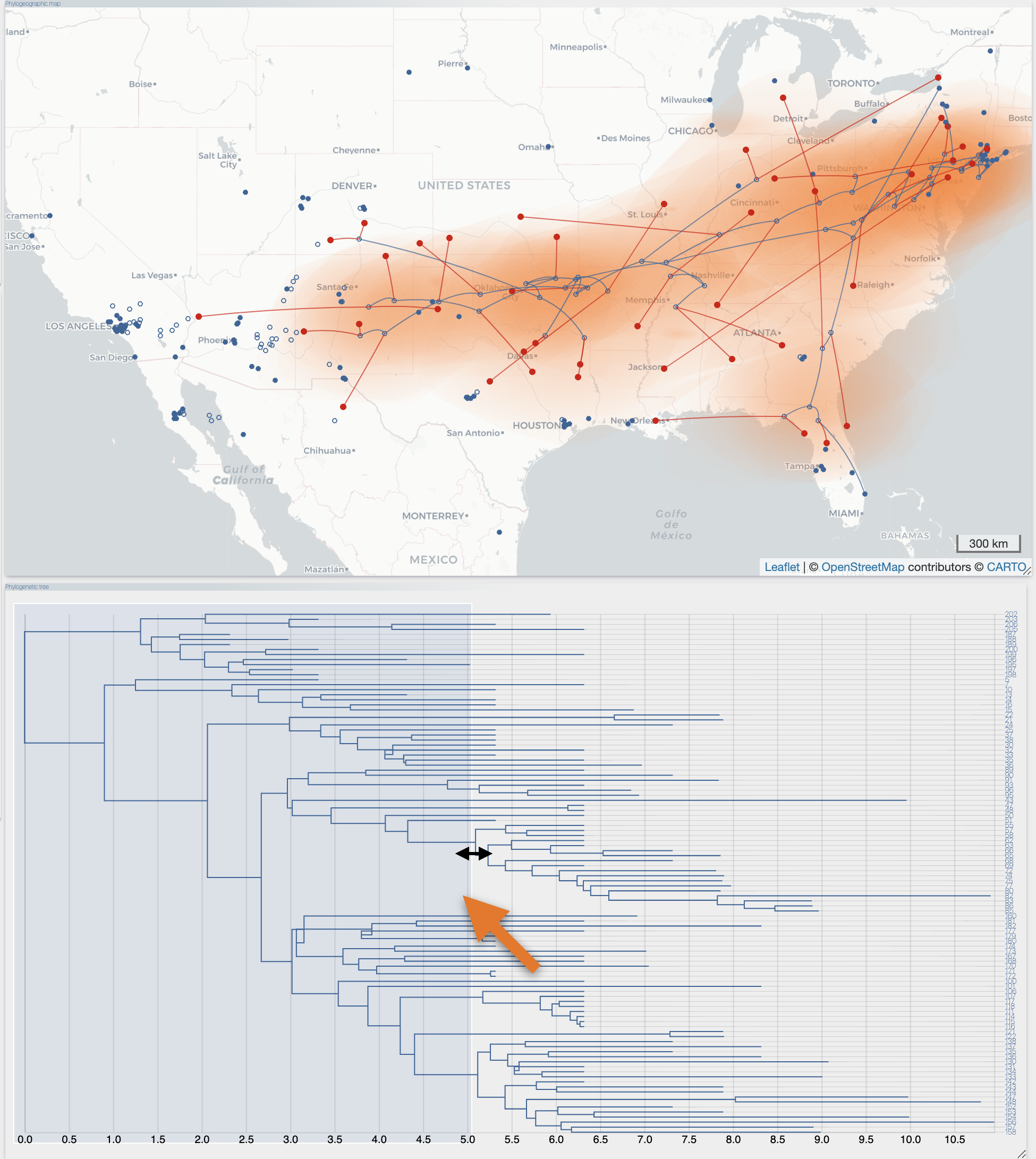
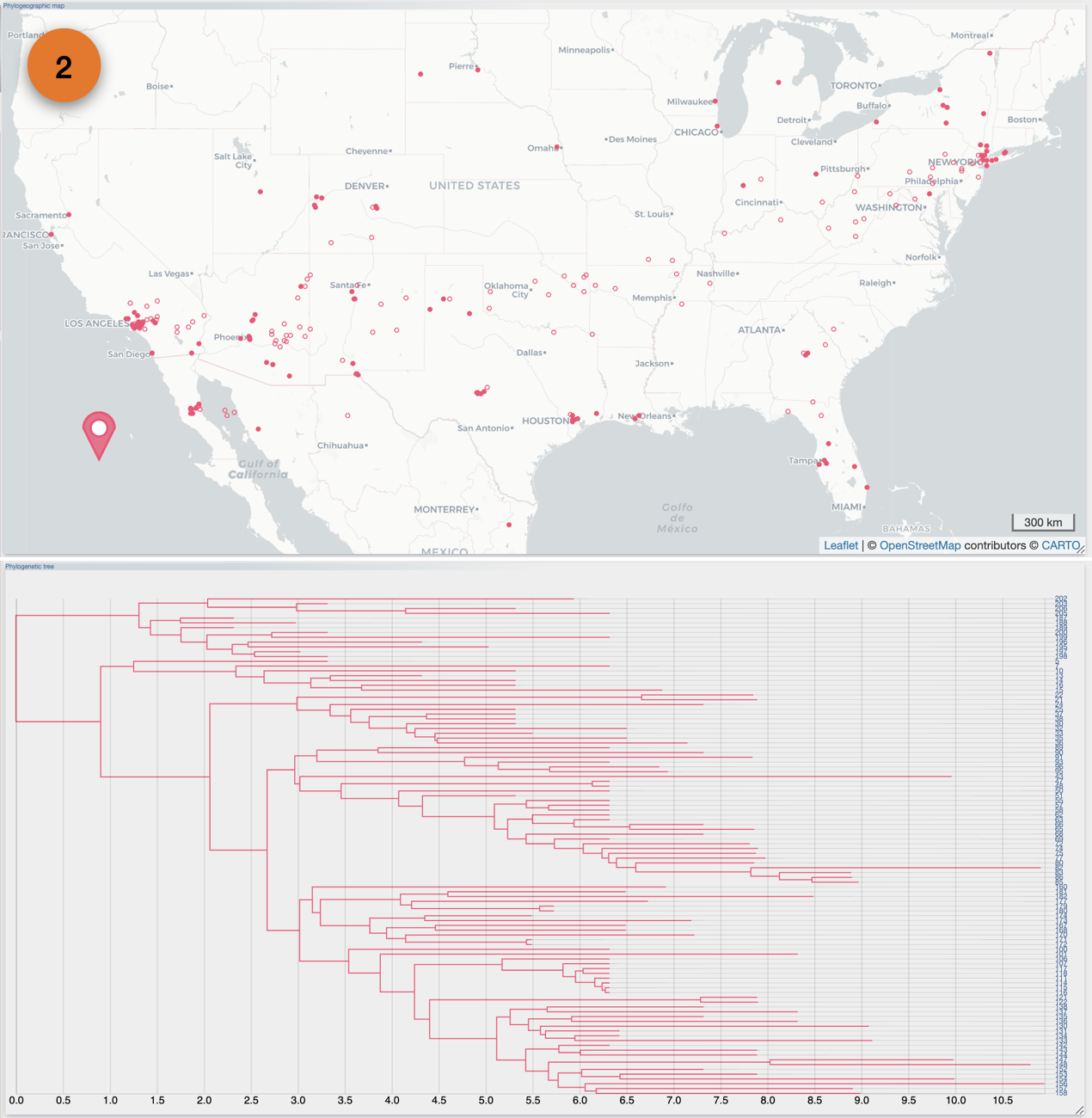
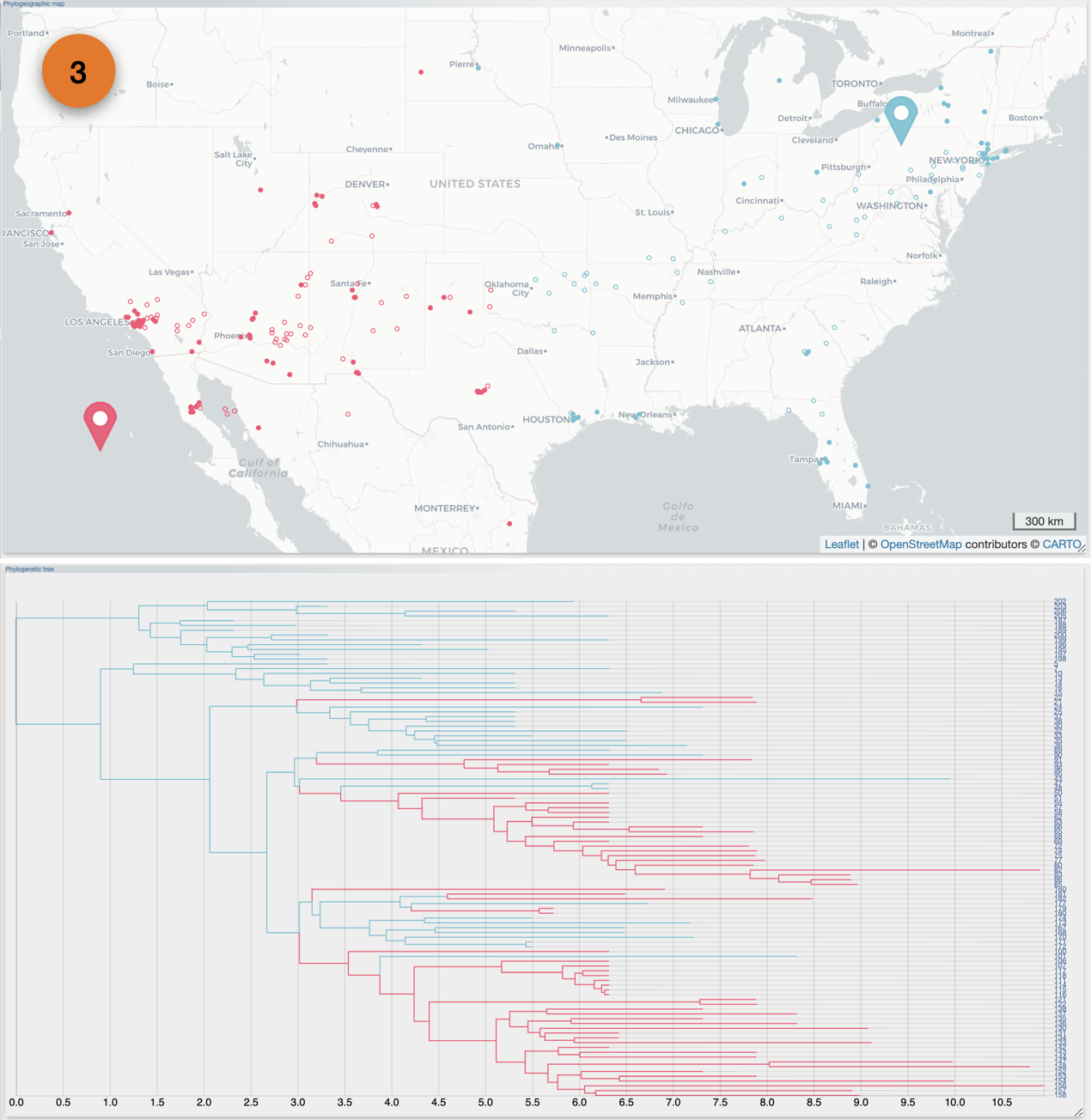
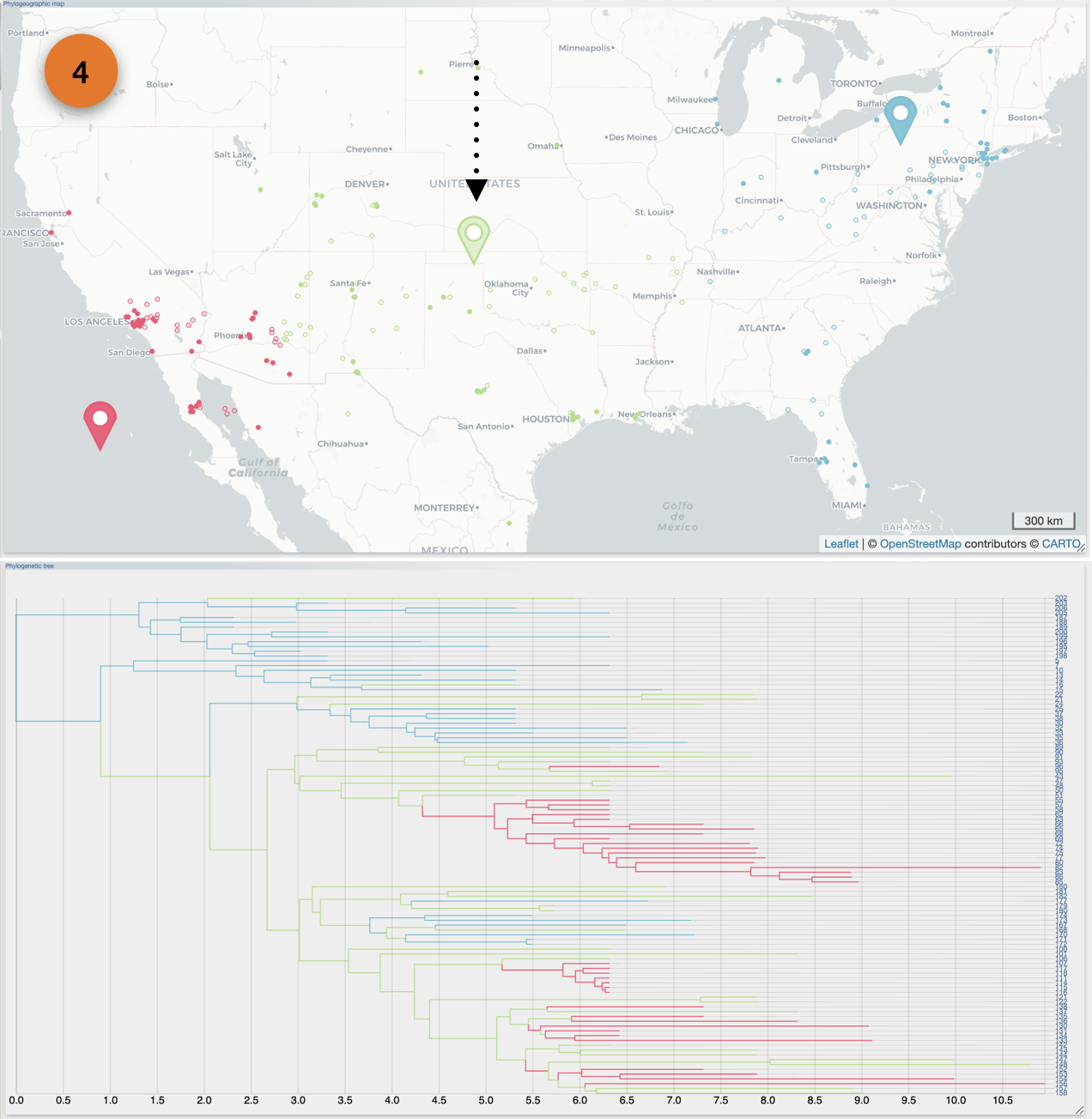
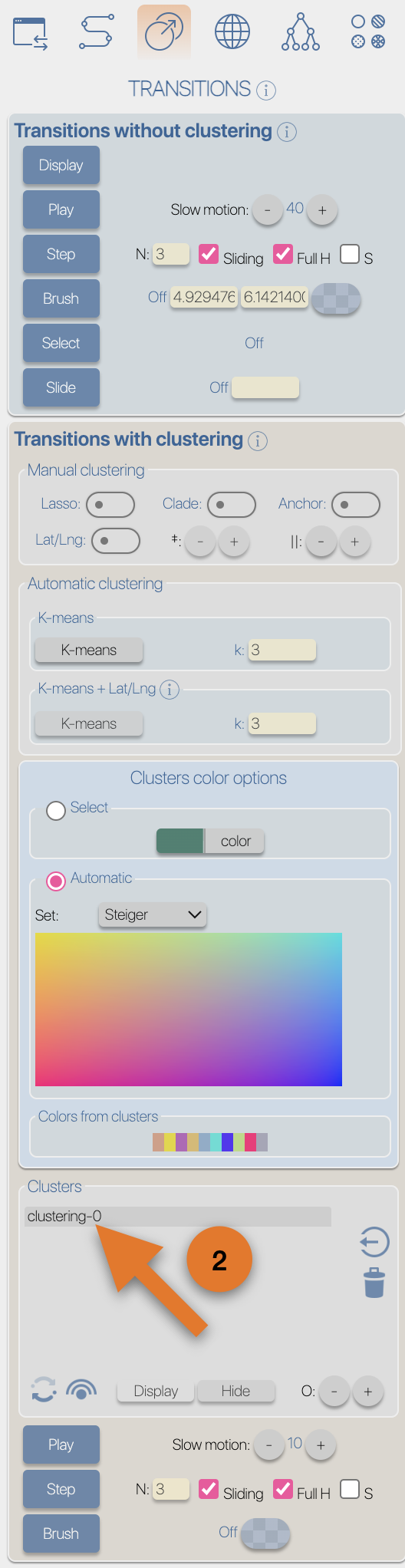
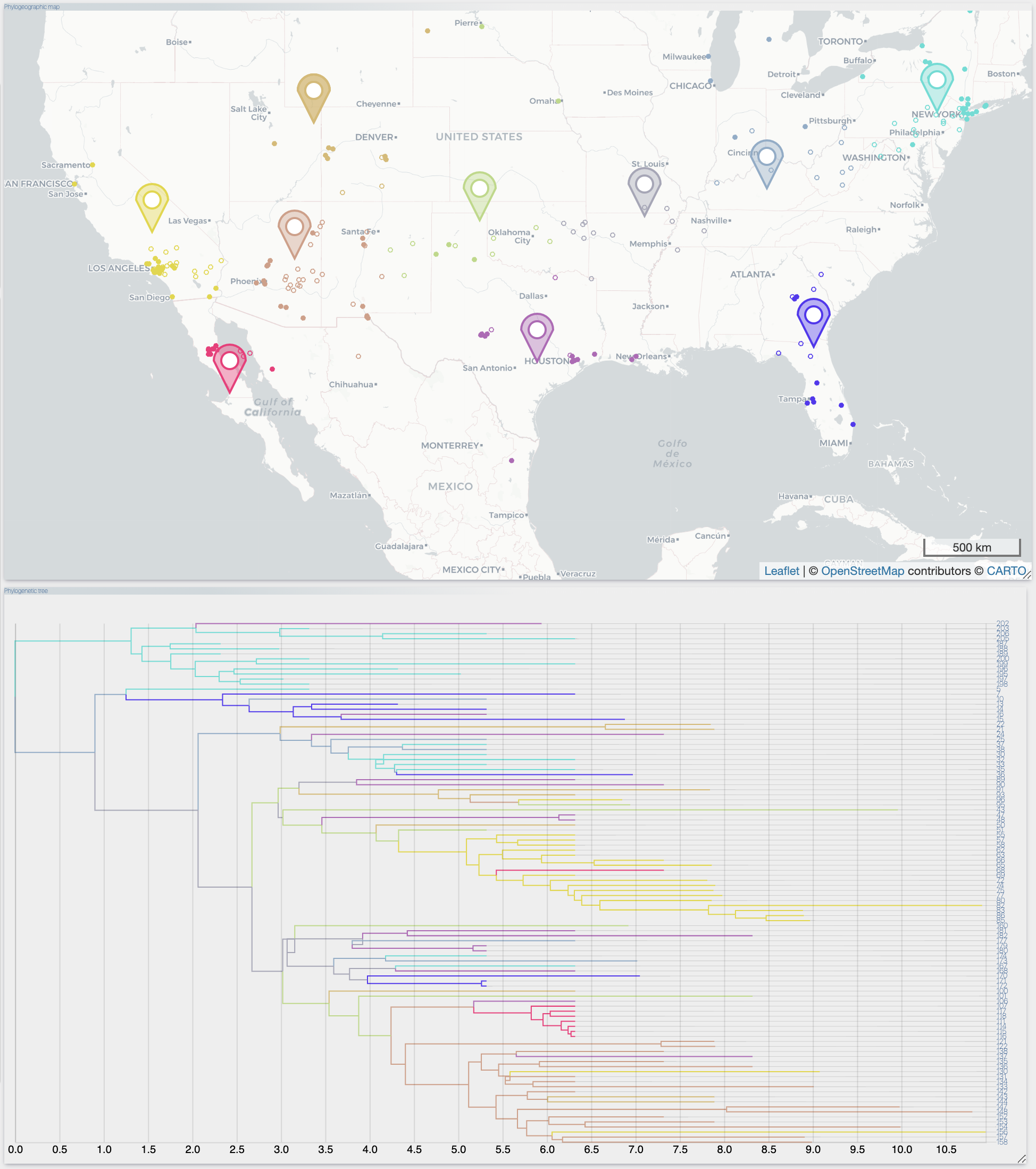
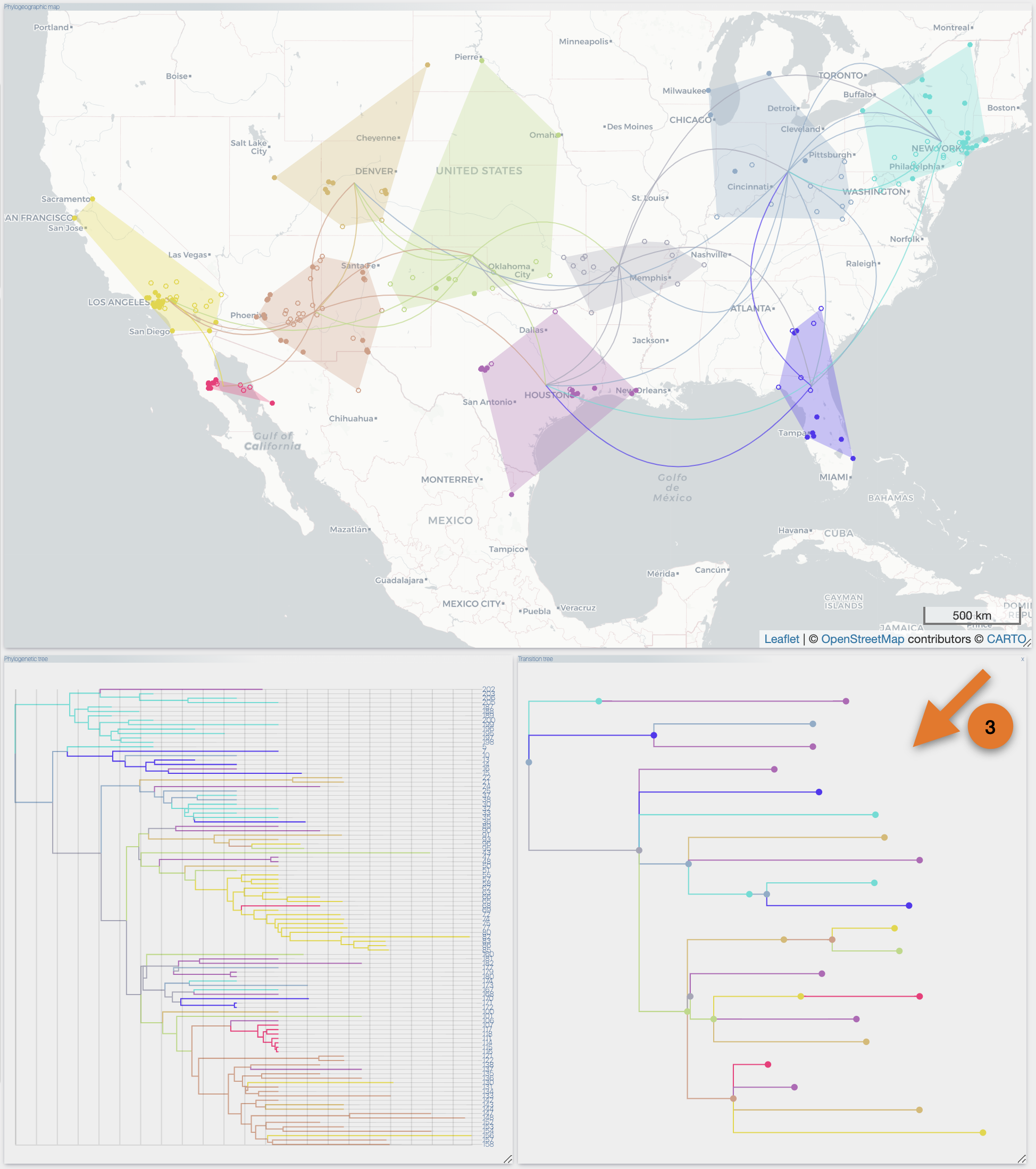
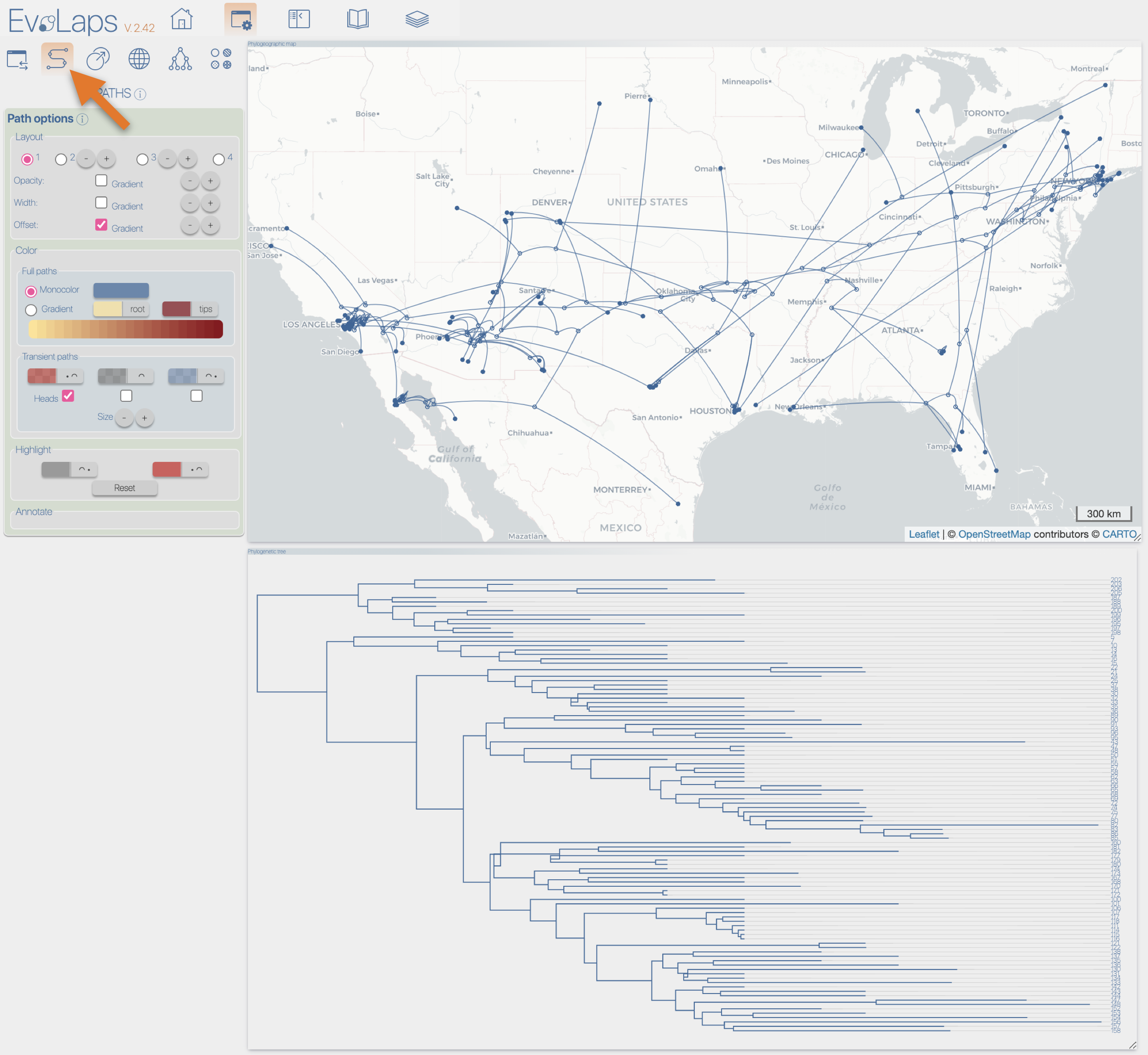
After the importation process, EvoLaps switch automatically to the 'PATHS' toolbox, and displays the geographic map with the phylogeographic scenario and the phylogenetic tree.
|
|
|