|
1
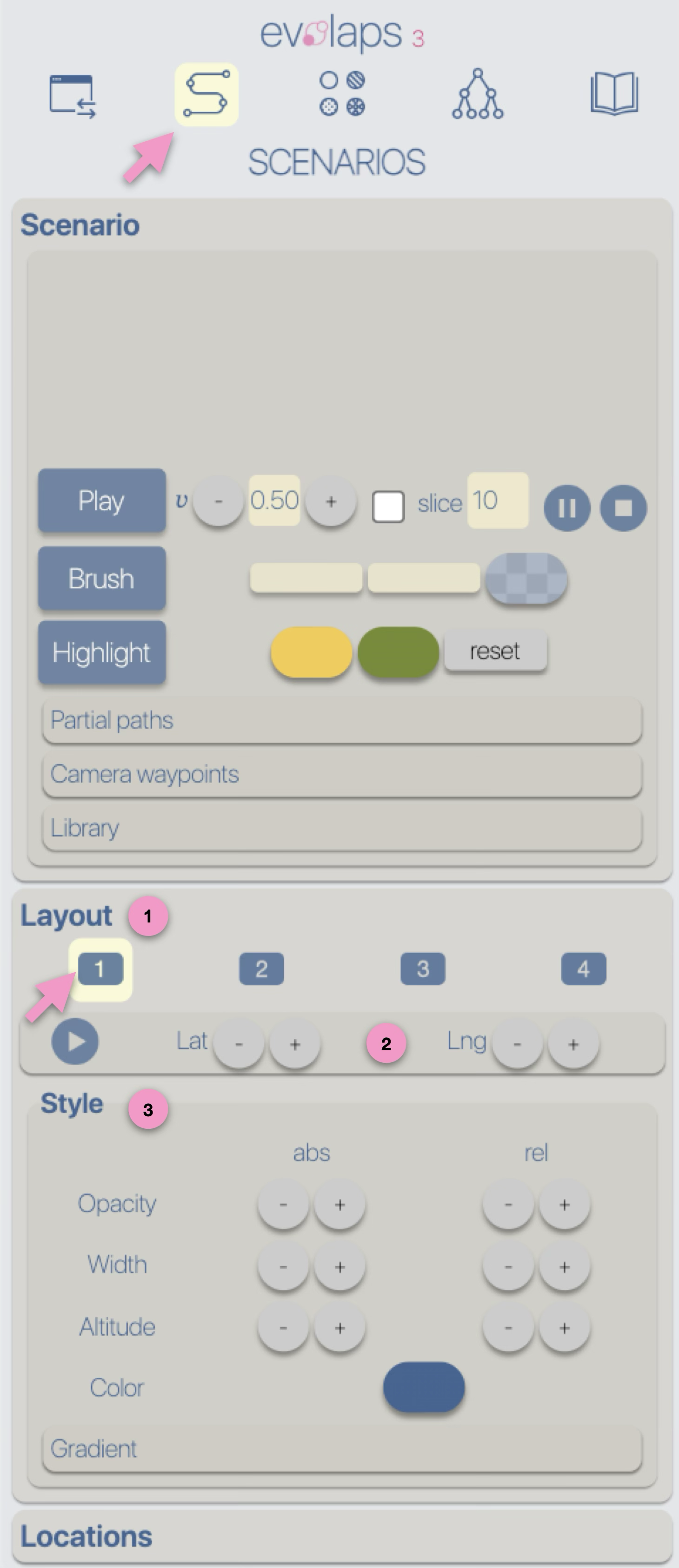
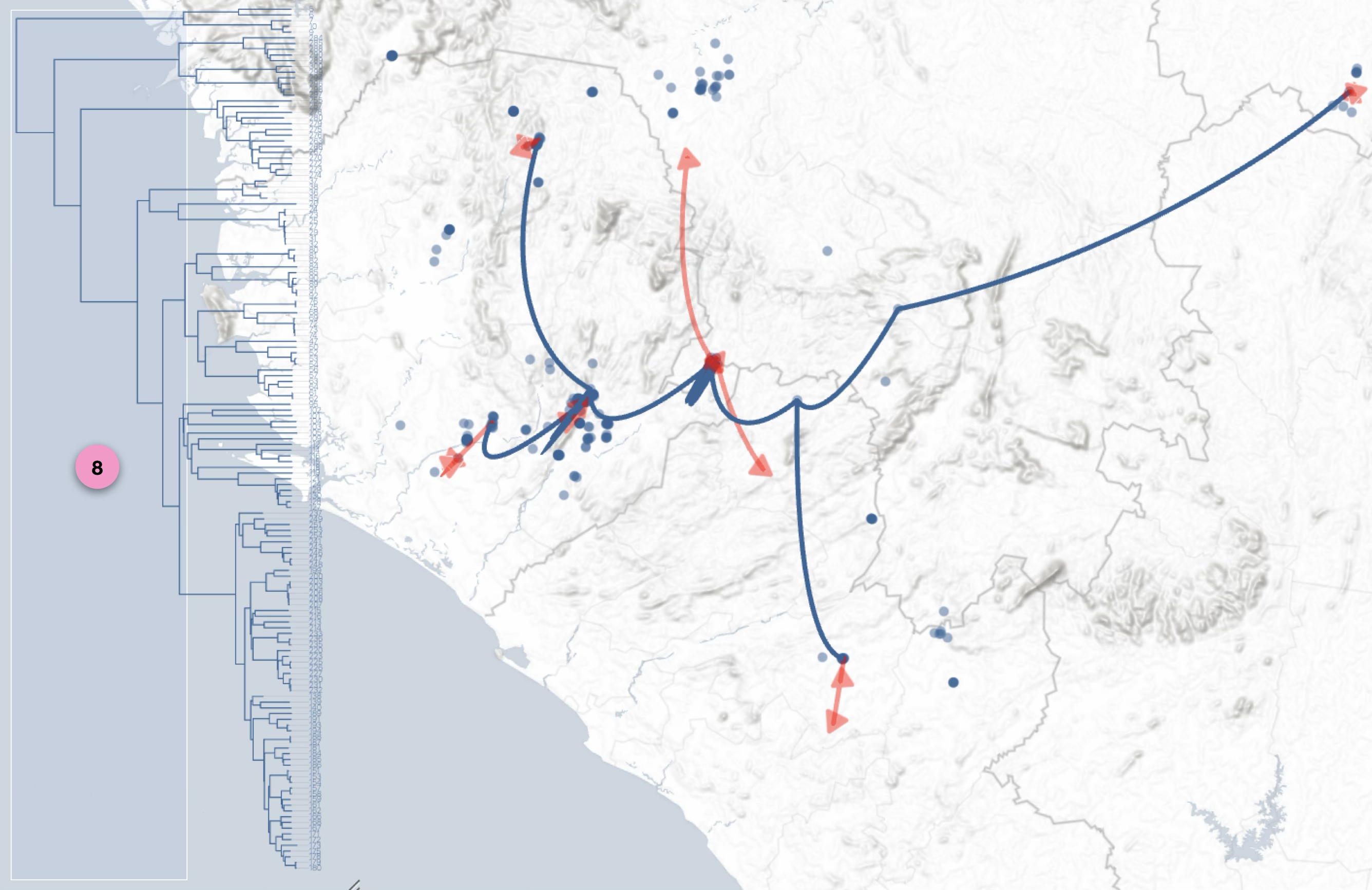
The graphical representations of phylogeographic scenarios (layouts) are grouped
into families (currently four), based on the methods used to compute a path connecting points (latitude/longitude)
associated with nodes of the phylogenetic tree (a parent node to its child nodes).
For example, and without going into too much technical detail, layout family 1 implements
a Bézier curve smoothing technique using three control points:
- a first point (latitude/longitude) associated with a parent node,
- a second point (latitude/longitude) associated with a child node,
- an intermediate point calculated as the average of the latitude and longitude of the two previous points.
for more details related to the layout families, see the Phylogeographic scenario > Layout section.
2
Layout-family-specific parameters allow for visual variations of the phylogeographic scenarios.
For instance, family 1, provides an offset control for the latitude and longitude of the intermediate point.
Nota Bene:
the graphical rendering quality of a phylogeographic scenario varies depending on the nature of the dataset,
the different families/methods used to compute the paths may be more or less suitable.
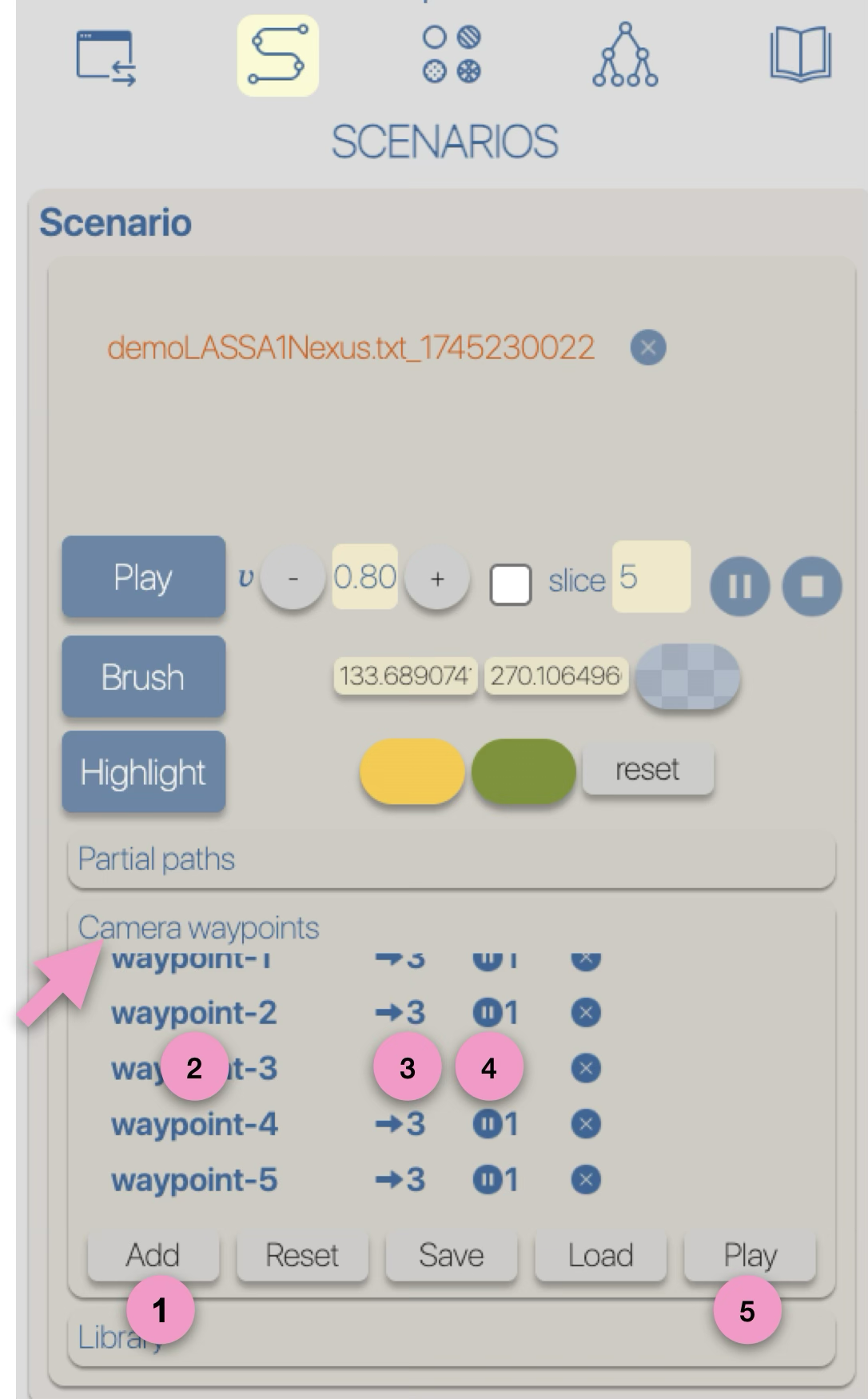
3
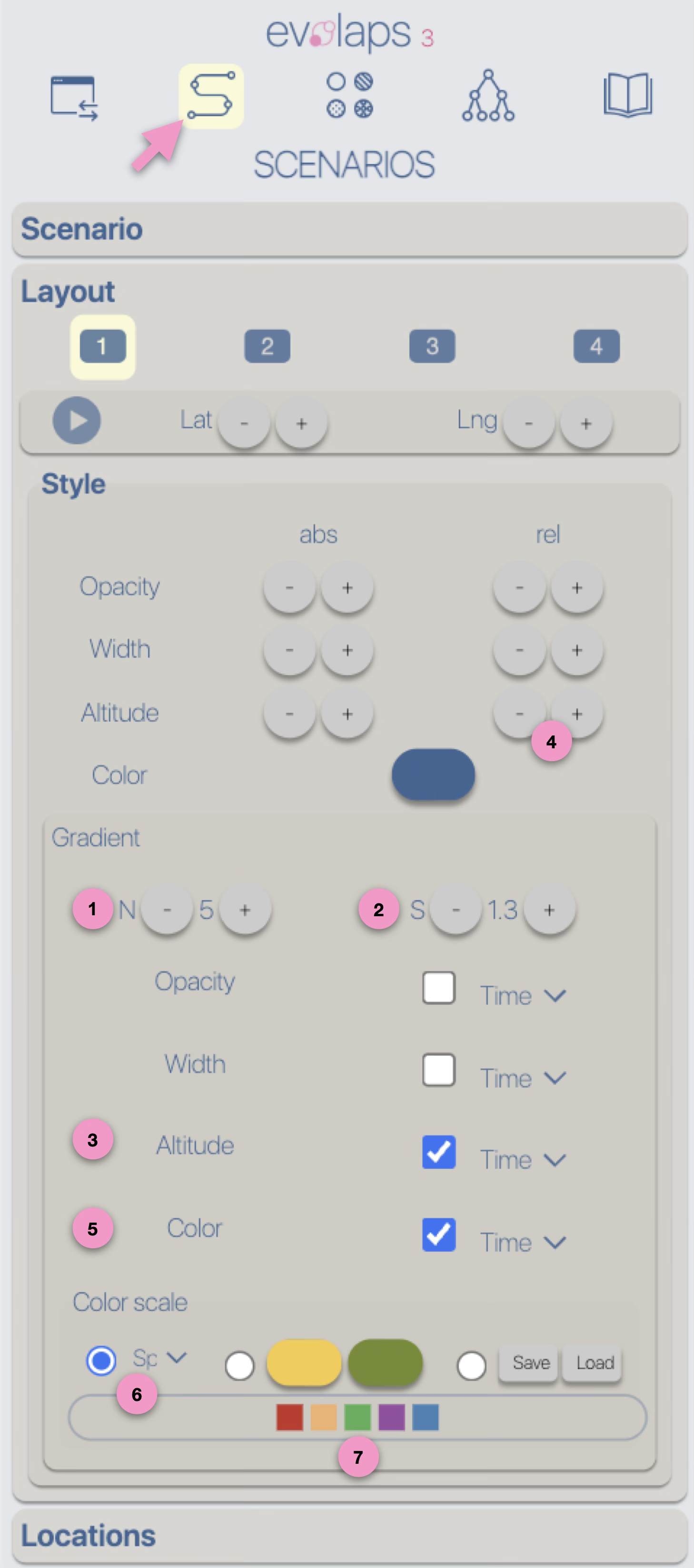
Style
Regardless of the layout used to represent the phylogeographic scenario, it can be modified using
four graphical variables: opacity, stroke thickness, altitude (curvature), and color.
Apart from color, modifications (increase/decrease) can be either absolute ('abs'): the same value is applied to all paths in the scenario, or
relative ('rel'): the existing value of each path is incremented (or decremented) individually.
These modifications can be performed manually (using -/+ buttons) or automatically by applying a gradient (next step of the tutorial).
for more details related to the style, see the Phylogeographic scenario > Style section.
|






 .
. .
.